Abstract
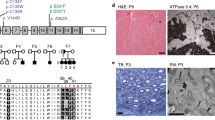
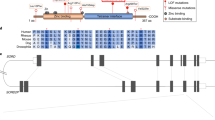
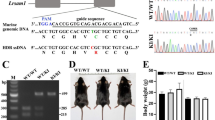
Hereditary sensory neuropathy type I (HSN1) is the most common hereditary disorder of peripheral sensory neurons. HSN1 is an autosomal dominant progressive degeneration of dorsal root ganglia and motor neurons with onset in the second or third decades. Initial symptoms are sensory loss in the feet followed by distal muscle wasting and weakness. Loss of pain sensation leads to chronic skin ulcers and distal amputations1,2. The HSN1 locus has been mapped to chromosome 9q22.1–22.3 (refs. 3,4). Here we map the gene SPTLC1, encoding serine palmitoyltransferase, long chain base subunit-1, to this locus. Mutation screening revealed 3 different missense mutations resulting in changes to 2 amino acids in all affected members of 11 HSN1 families. We found two mutations to be located in exon 5 (C133Y and C133W) and one mutation to be located in exon 6 of SPTLC1 (V144D). All families showing definite or probable linkage to chromosome 9 had mutations in these two exons. These mutations are associated with increased de novo glucosyl ceramide synthesis in lymphoblast cell lines in affected individuals. Increased de novo ceramide synthesis triggers apoptosis5,6 and is associated with massive cell death during neural tube closure7, raising the possibility that neural degeneration in HSN1 is due to ceramide-induced apoptotic cell death.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Dyck, P.J. Neuronal atrophy and degeneration predominantly affecting peripheral sensory and autonomic neurones. in Peripheral Neuropathy (eds. Dyck, P.J., Thomas, P.K., Griffin, J.W., Low, P.A. & Poduslo, J.F.) 1065–1093 (W B Saunders, Philadelphia, 1993).
Thomas, P.K. Hereditary sensory neuropathies. Brain Pathol. 3, 157–163 (1993).
Nicholson, G.A. et al. The gene for hereditary sensory neuropathy type I (HSN-I) maps to chromosome 9q22.1–q22.3. Nature Genet. 13, 101–104 (1996).
Bejaoui, K. et al. Confirmation of linkage of type 1 hereditary sensory neuropathy to human chromosome 9q22. Neurology 52, 510–515 (1999).
Perry, D.K. et al. Serine palmitoyltransferase regulates de novo ceramide generation during etoposide-induced apoptosis. J. Biol. Chem. 275, 9078–9084 (2000).
Shimabukuro, M. et al. Lipoapoptosis in β-cells of obese prediabetic fa/fa rats. J. Biol. Chem. 273, 32487–32490 (1998).
Herget, T. et al. Production of ceramides causes apoptosis during early neural differentiation in vitro. J. Biol. Chem. 275, 30344–30354 (2000).
Blair, I.P., Hulme, D., Dawkins, J.L. & Nicholson, G.A. A YAC-based transcript map of human chromosome 9q22.1–q22.3 encompassing the loci for hereditary sensory neuropathy type I and multiple self-healing squamous epithelioma. Genomics 51, 277–281 (1998).
Weiss, B. & Stoffel, W. Human and murine serine-palmitoyl-CoA transferase. Cloning, expression and characterization of the key enzyme in sphingolipid synthesis. Eur. J. Biochem. 249, 239–247 (1997).
Auer-Grumbach, M., Wagner, K., Timmerman, V., De Jonghe, P. & Hartung, H.P. Ulcero-mutilating neuropathy in an Austrian kinship without linkage to hereditary motor and sensory neuropathy IIB and hereditary sensory neuropathy I loci. Neurology 54, 45–52 (2000).
Nagiek, M.M., Lester, R.L. & Dickson, R.C. Sphingolipid synthesis: identification and characterization of mammalian cDNAs encoding the Lcb2 subunit of serine palmitoyltransferase. Gene 177, 237–241 (1996).
Mathews, C.K. & van Holde, K.E. in Biochemistry 617 (Benjamin/Cummings, Redwood City, 1990).
Snell, E.E., Dimari, S.J. & Brady, R.N. Biosynthesis of sphingosine and dihydrosphingosine by cell-free systems from Hansenula ciferri. Chem. Phys. Lipids 5, 116–138 (1970).
Medlock, K.A. & Merril, J.A.H. Inhibition of serine palmitoyltransferase in vitro and long-chain base biosynthesis in intact Chinese hamster ovary cells by β-chloroalanine. Biochemistry 27, 7079–7084 (1988).
Hanada, K. et al. A mammalian homolog of the yeast LCB1 encodes a component of serine palmitoyltransferase, the enzyme catalyzing the first step in sphingolipid synthesis. J. Biol. Chem. 272, 32108–32114 (1997).
Merrill, J.A.H. et al. Sphingolipids—the enigmatic lipid class: biochemistry, physiology, and pathophysiology. Toxicol. Appl. Pharmacol. 142, 208–225 (1997).
Stanley, C.A. et al. Hyperinsulinism and hyperammonemia in infants with regulatory mutations of the glutamate dehydrogenase gene. N. Engl. J. Med. 338, 1352–1357 (1998).
Andreutti-Zaugg, C., Scott, R.J. & Iggo, R. Inhibition of nonsense-mediated messenger RNA decay in clinical samples facilitates detection of human MSH2 mutations with an in vivo fusion protein assay and conventional techniques. Cancer Res. 57, 3288–3293 (1997).
Sarkar, G., Yoon, H.S. & Sommer, S.S. Dideoxy fingerprinting (ddF): a rapid and efficient screen for the presence of mutations. Genomics 13, 441–443 (1992).
Bligh, E.A. & Dyer, W.J. A rapid method for total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 (1959).
Acknowledgements
We thank the family members for participation; M. Jenkins for assistance with sample collections; T. Bananis for organizational assistance and cell culture work; K. Hanada and S. Samman for advice on the ceramide assay; M. Kennerson and D. Handelsman for critical review of the manuscript; M. Lloyd and B. Christie for assistance with photography; P.K. Thomas, M. Campbell, D. Ellison, N. Wood and P. Ashby for providing access to families; and M. Williams for assistance with blood collections and DNA preparations. This work was supported by grants from the National Health and Medical Research Council of Australia, and the Muscular Dystrophy Association of the USA. J. Dawkins was responsible for finding the mutations in SPTLC1 and preparation of the manuscript; D. Hulme found the raised glucosyl ceramide levels.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Rights and permissions
About this article
Cite this article
Dawkins, J., Hulme, D., Brahmbhatt, S. et al. Mutations in SPTLC1, encoding serine palmitoyltransferase, long chain base subunit-1, cause hereditary sensory neuropathy type I. Nat Genet 27, 309–312 (2001). https://doi.org/10.1038/85879
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/85879
This article is cited by
-
Deciphering lipid dysregulation in ALS: from mechanisms to translational medicine
Translational Neurodegeneration (2022)
-
Lipid and metabolic alteration involvement in physiotherapy for chronic nonspecific low back pain
Lipids in Health and Disease (2022)
-
Neurological update: hereditary neuropathies
Journal of Neurology (2022)
-
Kicking off sphingolipid biosynthesis: structures of the serine palmitoyltransferase complex
Nature Structural & Molecular Biology (2021)
-
Serine administration as a novel prophylactic approach to reduce the severity of acute pancreatitis during diabetes in mice
Diabetologia (2020)