Abstract
Aims/hypothesis
Genotype does not change over the life course and may thus facilitate earlier identification of individuals at high risk for type 2 diabetes. We hypothesised that a genotype score predicts incident type 2 diabetes from young adulthood and improves diabetes prediction models based on clinical risk factors alone.
Methods
The Coronary Artery Risk Development in Young Adults (CARDIA) study followed young adults (aged 18–30 years, mean age 25) serially into middle adulthood. We used Cox regression to build nested prediction models for incident type 2 diabetes based on clinical risk factors assessed in young adulthood (age, sex, race, parental history of diabetes, BMI, mean arterial pressure, fasting glucose, HDL-cholesterol and triacylglyercol), without and with a 38-variant genotype score. Models were compared with C statistics and continuous net reclassification improvement indices (NRI).
Results
Of 2,439 participants, 830 (34%) were black and 249 (10%) had a BMI ≥30 kg/m2 at baseline. Over a mean 23.9 years of follow-up, 215 (8.8%) participants developed type 2 diabetes. The genotype score significantly predicted incident diabetes in all models, with an HR of 1.08 per risk allele (95% CI 1.04, 1.13) in the full model. The addition of the score to the full model modestly improved reclassification (continuous NRI 0.285; 95% CI 0.126, 0.433) but not discrimination (C statistics 0.824 and 0.829 in full models with and without score). Race-stratified analyses were similar.
Conclusions/interpretation
Knowledge of genotype predicts type 2 diabetes over 25 years in white and black young adults but may not improve prediction over routine clinical measurements.
Similar content being viewed by others
Introduction
Type 2 diabetes can be delayed or prevented with lifestyle modification [1–3]. If clinicians could identify type 2 diabetes risk earlier in life, they might reduce lifetime morbidity through targeted prevention in high-risk individuals. Young adulthood might be the opportune time to make an early intervention on any health-related behaviour that might predispose an individual to type 2 diabetes later in life. Of the many prediction models for incident type 2 diabetes in adulthood, few have included adequate numbers of young adults [4]. In addition, type 2 diabetes prediction models developed in cohorts of middle-aged adults may have inferior performance in younger adults [5].
Type 2 diabetes is heritable [6, 7]. Large genome-wide association studies have identified at least 40 single-nucleotide polymorphisms (SNPs) independently associated with the disease, mostly among individuals of European ancestry [8–10]. The physiology underlying the relationship between most of these SNPs and type 2 diabetes is unknown. Genotype scores consisting of combinations of these SNPs predict incident type 2 diabetes in middle-aged adults but do not meaningfully improve prediction models consisting of traditional risk factors such as family history and BMI [11–13] . It is unknown whether such a genotype score might improve type 2 diabetes prediction from young adulthood. Young adults may not yet fully demonstrate clinically identifiable risk factors, such as overweight/obesity, dyslipidaemia, or family history, which would allow clinicians to assess their future diabetes risk. Moreover, no prospective study has assessed whether a genotype score predicts incident type 2 diabetes exclusively among individuals of African ancestry. We used data from the Coronary Artery Risk Development in Young Adults (CARDIA) study to test whether the addition of a genotype score improves type 2 diabetes prediction from young adulthood in whites and blacks.
Methods
Study design
The CARDIA study is a multicentre prospective study of 5,115 white and black individuals recruited from four cities across the USA in 1985–1986 [14, 15]. Study participants were 18–30 years old at the baseline examination and were invited to participate in serial follow-up examinations and surveys up to 25 years after the initial study visit. Retention of the original cohort was 72% at the year 25 examination. Informed consent was obtained from all participants, and the institutional review board at each participating centre approved the study.
Inclusion and exclusion criteria
We limited the present analyses to unrelated study participants who attended at least one follow-up examination and for whom genotype information and baseline data were available for all predictors of interest. We excluded any participant with diabetes or pregnancy at the baseline examination. Participants who reported diabetes treatment exclusively with insulin during the observation period were considered to have type 1 diabetes and were also excluded from analyses.
Incident type 2 diabetes
We identified cases of type 2 diabetes according to the World Health Organization definition [16]: fasting plasma glucose ≥7.0 mmol/l (≥126 mg/dl) or report of taking oral diabetes medications with or without insulin injections. Time to event was calculated from the date of the baseline examination to the date of the first follow-up examination meeting our criteria for incident type 2 diabetes (cases) or to the date of the last CARDIA examination for each participant without incident type 2 diabetes (censored individuals).
Measurement of covariates
Data collection protocols have been described previously [15]. The baseline survey in 1985–1986 assessed self-reported race (black or white) and parental history of diabetes; it did not specify diabetes type. Weight, height, systolic blood pressure (SBP) and diastolic blood pressure (DBP) were measured using standard protocols with participants wearing light clothing and no shoes. BMI was calculated as weight (kg) divided by height (m) squared. Mean arterial pressure (MAP) was calculated as 1/3(SBP) + 2/3(DBP). Morning venous blood samples were obtained after an overnight fast of at least 8 h. Fasting plasma glucose and insulin were measured with the hexokinase–ultraviolet and radioimmunoassay methods, respectively (Linco Research, St Charles, MO, USA). The details of the measurement of HDL-cholesterol and triacylglyercol have been reported previously [17]. Baseline smoking status, alcohol intake and tertile of physical activity were determined as previously reported [18].
Genotyping and genotype risk score
CARDIA samples were genotyped with the Affymetrix Genome-Wide Human SNP Array 6.0 (Santa Clara, CA, USA) as described previously [19, 20]. To calculate the genotype score, we used 38 SNPs associated with type 2 diabetes at the genome-wide significant level (see electronic supplementary material [ESM] Table 1). The score was the unweighted sum of the number of risk alleles (0, 1 or 2) at each of the 38 SNP loci. Genotyped or imputed data were available for the 38 SNPs in all white participants, but no genotyped or imputed value for rs757210 was available for any black participant. Black individuals were thus given a score of 1 for that missing value, equivalent to being heterozygous at that locus, the most common genotype at those loci among whites. Details of the imputation and quality of CARDIA genotyping have been published previously [19, 20]. Principal components used for supplemental analyses of ancestry were calculated as previously described [20].
Statistical analyses
We used Cox proportional hazards regression to build nested models for time to incident type 2 diabetes. Models sequentially included: (1) demographics (age, sex and self-reported race); (2) parental history of diabetes (yes vs no); (3) physical examination (BMI and MAP) and (4) routine laboratory predictors (fasting glucose, natural log-transformed HDL-cholesterol, and natural log-transformed triacylglyercol)—all as assessed at the baseline examination. We based this prediction model on a clinical prediction model validated in the Framingham Offspring Study [21]. Ties were handled with the exact method. To test the validity of the proportional hazards assumption, we included a time-dependent interaction term with each variable in every model. We rejected the proportional hazards assumption for a given variable if its interaction term with time had p < 0.05. All variables met the proportional hazards assumption in every model unless otherwise stated. In black participants, sex did not meet the proportional hazards assumption, and thus models in blacks are sex-stratified. If a continuous predictor did not meet the proportional hazards assumption in a model over the entire follow-up period, an HR was calculated as the highest integer value of follow-up time in years for which it did meet the proportional hazards assumption (see footnotes in Tables 2 and 3) [22]. We assessed model calibration with a Hosmer–Lemeshow χ 2 statistic comparing observed and expected event counts in deciles of event probability [23].
The 38-SNP genotype score was then added to each of these models, and model improvement was assessed. The fit of nested models was compared with likelihood ratio tests. We calculated C statistics and continuous net classification improvement (NRI) indices for survival data and their 95% CIs using the methods described by Pencina et al [24, 25]. Unlike categorical NRI indices, the continuous NRI does not require discrete risk categories and relies, rather, on the proportions of cases correctly assigned a higher model probability and non-cases correctly assigned a lower model probability by a second model compared with the first [25]. Continuous NRI values of 0.2 correspond to weak improvement in reclassification, while values of 0.4 correspond to an intermediate effect [26]. Hosmer–Lemeshow statistics, C statistics and NRI were calculated at 25 years of follow-up. We estimated 95% CIs around the NRI using 999 bootstrap replications [25, 27].
We performed the above analyses in the overall cohort and in each race separately. To identify potential differences by parental history or BMI in the effect of genotype score on diabetes risk, we tested the statistical significance of interaction terms in the full model: parental history (yes vs no) by genotype score and BMI (<25 vs ≥25 kg/m2) by genotype score. One set of supplemental models additionally included baseline smoking status, alcohol consumption and tertile of physical activity and a second set included the first two principal components of ancestry in the model instead of self-reported race. To compare risks associated with continuous variables (age, BMI, MAP, glucose, log-transformed HDL-cholesterol, log-transformed triacylglyercol and genotype score), we additionally calculated standardised HRs corresponding to the increased hazard associated with a 1-SD increase in each variable. All analyses were performed with SAS v. 9.3 software (SAS Institute, Cary, NC, USA).
Results
Baseline characteristics
Of 2,439 eligible individuals, 830 (34%) were black and 1,364 (56%) were women (Table 1). At baseline, 6.5% of the white and 17.4% of the black participants had a BMI ≥ 30 kg/m2. On average, each participant had 41 risk alleles across the 38 loci, with blacks having significantly more than whites (42.6 vs 40.2, p < 0.001).
Incident type 2 diabetes
Over a mean follow-up of 23.9 (SD 2.8) years, 215 (8.8%) participants developed type 2 diabetes. The incidence rate of type 2 diabetes was 3.8, 2.5 and 6.4 per 1,000 person-years in the overall cohort, in white participants and in black participants, respectively. The mean age at type 2 diabetes detection was 44.1 (SD 6.6) years.
Genotype and incident type 2 diabetes
After adjustment for age, sex and race, each additional risk allele of the 38-SNP genotype score was associated with a 9% (95% CI 5, 13) greater hazard of incident type 2 diabetes over the study period (p < 0.001). Each one-SD increase in genotype score, BMI and fasting glucose was associated with an HR of 1.56 (95% CI 1.36, 1.79), 1.98 (95% CI 1.83, 2.15), and 1.54 (95% CI 1.36, 1.75), respectively, in age- and sex-adjusted models. Genotype score remained a significant predictor of incident type 2 diabetes in all sequentially more complex prediction models (all p < 0.05; Table 2). After adjustment for demographics, parental history, physical examination and laboratory analyses, genotype score had an HR of 1.08 (95% CI 1.04, 1.13) for each additional risk allele.
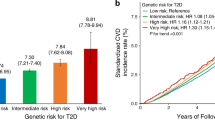
In the fully adjusted model, those in the highest tertile of genotype score (43–53 risk alleles) had an HR of 1.77 (95% CI 1.23, 2.54) for type 2 diabetes compared with the lowest tertile (29–39 risk alleles). In contrast, the tertile with the highest BMI (25–53 kg/m2) had an HR of 4.37 (95% CI 2.68, 7.12) compared with the lowest tertile (14–22 kg/m2; Fig. 1). When the fully adjusted analyses were stratified by baseline BMI <25 or ≥25 kg/m2, each one-allele increase in genotype score had an HR of 1.07 (95% CI 1.02, 1.11) in those with BMI ≥25 kg/m2 and 1.13 (95% CI 1.05, 1.21) in those with BMI <25 kg/m2. This difference was significant when tested as an interaction term in the fully adjusted model in the overall cohort (p < 0.001) but not in each racial group analysed separately (p > 0.05). The effect of genotype score did not differ among those with and without a parental history of diabetes (p > 0.05 for interaction term).
Comparison of prediction models without and with genotype score
The addition of genotype score did not significantly increase the C statistics of any of the four sequentially nested clinical prediction models, although the absolute increase in the C statistics of the models with demographics or with demographics and parental history suggested improved discrimination (Table 2). As assessed by the continuous NRI, genotype score resulted in modest improvement in model reclassification (NRI range 0.206–0.285) (Table 2). Neither the addition of baseline smoking status, alcohol consumption and tertile of physical activity (ESM Tables 2 and 3) nor the inclusion of ancestry principal components instead of self-reported race (ESM Table 4) changed these results.
Race-stratified analyses
When white and black participants were analysed separately, genotype score was associated with incident type 2 diabetes in all models in both racial groups (Table 3). In the full model, each additional risk allele had an HR of 1.07 (95% CI 1.01, 1.13) in whites and 1.09 (95% CI 1.04, 1.15) in blacks. In both whites and blacks, the absolute increases in C statistics indicated improvement in model discrimination with the addition of genotype score to models consisting of demographics and parental history but no improvement in more complex models. The addition of genotype score achieved modest improvement in reclassification, which did not appear to differ by race.
Discussion
The early identification of individuals at risk for future type 2 diabetes may enable targeted prevention efforts. We have shown that genotype information, combined as an aggregate risk score, predicts incident type 2 diabetes over 25 years of follow-up from young adulthood. Genotype score remained a significant predictor after adjustment for other type 2 diabetes risk factors routinely assessed in clinical care, including parental history. Although most of the variants comprising the 38-variant genotype score were discovered in populations of predominantly European ancestry, the score was a significant predictor of diabetes risk among both white and black young adults.
The addition of the genotype score to a full clinical prediction model for type 2 diabetes did not, however, significantly increase model C statistic. Because the C statistic may not be a suitable assessment of a prediction model’s clinical utility [28], we also calculated continuous NRI indices comparing prediction models with and without the genotype score. The addition of genotype score to the full clinical prediction model resulted in an NRI of 0.285, indicating modest improvement in model reclassification [26].
A few prospective studies have demonstrated that genotype risk scores predict incident type 2 diabetes, but most of these studies have consisted only of middle-aged or older adults of European ancestry, followed on average for 10 years or less [11–13, 29–32]. In general, these studies have shown that genotype scores do not improve prediction models consisting of routine clinical risk factors including family history, BMI and fasting glucose. One exception, the Malmö Preventive Project, showed that the addition of an 11-variant genotype score to clinical prediction models for type 2 diabetes significantly improved model discrimination over a median 24.8 years of follow-up among middle-aged Swedish adults [11]. In that study, the performance of a genetic prediction model improved with increasing duration of follow-up relative to a clinical prediction model. Moreover, stratified analyses from the Framingham Offspring Study have demonstrated that genotype information may significantly improve diabetes risk reclassification in adults younger than 50 years old but not older [13]. Outside these data, young adulthood has not been a focus of the application of genotype information to type 2 diabetes risk prediction. The present findings in young adults are consistent with prior evidence in older adults and demonstrate that genotype information may achieve modest improvement over clinical prediction models for type 2 diabetes over 25 years of follow-up from young adulthood. We hypothesised that genotype information in young adulthood might have greater predictive value over clinical risk predictors than it does later in life. Genotype does not change over the life course, while routine risk factors for type 2 diabetes, including overweight, dyslipidaemia, elevated fasting glucose and even parental history of diabetes, may not manifest until later in adulthood. However, our results do not support the idea that genotype testing earlier in adulthood results in clinically meaningful improvement in type 2 diabetes risk prediction over the risk factors already measured in clinical practice.
A reasonable next question, then, is whether genotype information might improve type 2 diabetes prediction even earlier in the life course, before young adulthood. In a cohort of white and black adolescents followed for an average of 26 years into adulthood, we have found that a 38-SNP score also predicts incident type 2 diabetes [33]. Similarly, however, this score did not improve prediction over other clinical risk factors measured at baseline between ages 12 and 18 years, as assessed by the continuous NRI. Nonetheless, the predictive accuracy of these genotype models may be a conservative estimate, if certain of the individual loci represented in our summary SNP score exert their effects on diabetes risk at different ages. The present analyses would not detect such genotype-by-age interaction effects. Indeed, although this study was not powered to detect the small effect of each SNP individually, 13 of the 38 individual SNP effects were in the opposite direction from the published associations (ESM Table 1). Studies in larger cohorts with the power to model age-varying effects of individual risk variants and summary scores should be undertaken.
No previous study has reported a prospective association between genotype score and incident type 2 diabetes in a cohort of exclusively African-Americans. In a case–control study of middle-aged and older African-American adults, Palmer recently showed that each additional risk allele in an unweighted 17-SNP score was associated with an OR of 1.04 (95% CI 1.01, 1.08) for prevalent type 2 diabetes [34]. Although most type 2 diabetes-related SNPs were discovered in genome-wide association studies consisting predominantly of individuals of European ancestry, emerging evidence suggests that these loci, if not the specific SNPs themselves, are associated with type 2 diabetes risk in other ancestral groups [20, 35]. Our results suggest that a genotype score consisting of risk SNPs discovered in European cohorts may predict incident type 2 diabetes equally well in African-Americans.
Our study has certain limitations. First, the follow-up duration of 25 years is a strength of the present analyses, yet it may not be sufficiently long to allow accrual of an adequate number of cases of type 2 diabetes among those aged 18–30 years, now followed only to ages 43–55. A greater number of cases of type 2 diabetes will develop in the CARDIA cohort after even longer follow-up and may further strengthen the association between genotype score and diabetes risk. Second, our genotype score included common SNP variants associated with type 2 diabetes at the genome-wide significant level. The majority of these SNPs are not believed to be causal mutations and, indeed, are in intronic or intergenic regions of the genome [36]. As sequencing efforts identify the causal mutations for which most of these 38 SNPs are imperfect proxies, the incorporation of updated genetic information into such a risk score should strengthen its association with type 2 diabetes. Third, we summed all risk alleles across the 38 loci to calculate our genotype risk score, not accounting for the different effect size that each locus has with type 2 diabetes risk. While previous efforts have used genotype scores weighted for these different effect sizes [12, 13], we did not because the effect sizes between the majority of these loci and type 2 diabetes risk in blacks is unknown. Nonetheless, we demonstrated a significant association between the genotype score and type 2 diabetes, even after adjustment for clinical risk factors.
It remains unclear what threshold genotype information must reach to be clinically useful for type 2 diabetes prediction and prevention. For the prevention of CHD, a clinician can use the discrete 10-year risk categories of the Framingham Risk Score [37] to inform the LDL-cholesterol target and aspirin administration for an individual patient. No such paradigm exists for type 2 diabetes prevention. Moreover, the maintenance of a healthy weight, the mainstay of type 2 diabetes prevention, is advisable for everyone regardless of genotype [38]. However, the targeting of more resource-intensive preventive lifestyle modification programmes only to individuals at greatest risk might be warranted in a healthcare environment of finite resources. Still, our results do not conclusively demonstrate that genotype is the most appropriate measurement of that risk, even among young adults who may not yet have fully manifested their ultimate clinically measurable risk phenotypes.
In summary, a genotype risk score consisting of 38 common SNPs predicts incident type 2 diabetes among whites and blacks from young adulthood into the fifth and sixth decades of life. Young adulthood is an important period for type 2 diabetes prevention; during this time there is the potential to modify health-related behaviours that may influence risk later in life. At present, however, it is unclear whether genotype gives clinically actionable information for young adults.
Abbreviations
- CARDIA:
-
Coronary Artery Risk Development in Young Adults
- DBP:
-
Diastolic blood pressure
- MAP:
-
Mean arterial pressure
- NRI:
-
Net reclassification improvement index
- SBP:
-
Systolic blood pressure
- SNP:
-
Single-nucleotide polymorphism
References
Pan XR, Li GW, Hu YH et al (1997) Effects of diet and exercise in preventing NIDDM in people with impaired glucose tolerance. The Da Qing IGT and Diabetes Study. Diabetes Care 20:537–544
Knowler WC, Barrett-Connor E, Fowler SE et al (2002) Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Eng J Med 346:393–403
Tuomilehto J, Lindstrom J, Eriksson JG et al (2001) Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N Eng J Med 344:1343–1350
Noble D, Mathur R, Dent T, Meads C, Greenhalgh T (2011) Risk models and scores for type 2 diabetes: systematic review. BMJ 343:d7163
Mainous AG, Diaz VA, Everett CJ (2007) Assessing risk for development of diabetes in young adults. Ann Fam Med 5:425–429
Newman B, Selby JV, King MC, Slemenda C, Fabsitz R, Friedman GD (1987) Concordance for type 2 (non-insulin-dependent) diabetes mellitus in male twins. Diabetologia 30:763–768
Kaprio J, Tuomilehto J, Koskenvuo M et al (1992) Concordance for type 1 (insulin-dependent) and type 2 (non-insulin-dependent) diabetes mellitus in a population-based cohort of twins in Finland. Diabetologia 35:1060–1067
Diabetes Genetics Initiative of Broad Institute of Harvard MIT Lund University and Novartis Institutes of BioMedical Research, Saxena R, Voight BF et al (2007) Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316:1331–1336
Zeggini E, Scott LJ, Saxena R et al (2008) Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 40:638–645
Voight BF, Scott LJ, Steinthorsdottir V et al (2010) Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 42:579–589
Lyssenko V, Jonsson A, Almgren P et al (2008) Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Eng J Med 359:2220–2232
Meigs JB, Shrader P, Sullivan LM et al (2008) Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Eng J Med 359:2208–2219
de Miguel-Yanes JM, Shrader P, Pencina MJ et al (2011) Genetic risk reclassification for type 2 diabetes by age below or above 50 years using 40 type 2 diabetes risk single nucleotide polymorphisms. Diabetes Care 34:121–125
Cutter GR, Burke GL, Dyer AR et al (1991) Cardiovascular risk factors in young adults: the CARDIA baseline monograph. Control Clin Trials 12:1S–77S
Friedman GD, Cutter GR, Donahue RP et al (1988) CARIDA: study design, recruitment, and some characteristics of the examined subjects. J Clin Epidemiol 41:1105–1116
World Health Organization (1999) Part 1: Diagnosis and Classification of Diabetes Mellitus. In: World Health Organization Department of Noncommunicable Disease Surveillance (ed) Definition, diagnosis and classification of diabetes mellitus and its complications. Geneva, Switzerland
Bild DE, Jacobs DR Jr, Liu K et al (1996) Seven-year trends in plasma low-density-lipoprotein-cholesterol in young adults: the CARDIA Study. Ann Epidemiol 6:235–245
Hankinson AL, Daviglus ML, Bouchard C et al (2010) Maintaining a high physical activity level over 20 years and weight gain. JAMA 304:2603–2610
Lemaitre RN, Tanaka T, Tang W et al (2011) Genetic loci associated with plasma phospholipid n-3 fatty acids: a meta-analysis of genome-wide association studies from the CHARGE Consortium. PLoS Genet 7:e1002193
Lettre G, Palmer CD, Young T et al (2011) Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe Project. PLoS Genet 7:e1001300
Wilson PWF, Meigs JB, Sullivan L, Fox CS, Nathan DM, D’Agostino RB Sr (2007) Prediction of incident diabetes mellitus in middle-aged adults: the Framingham Offspring Study. Arch Intern Med 167:1068–1074
Franks PW, Hanson RL, Knowler WC et al (2007) Childhood predictors of young-onset type 2 diabetes. Diabetes 56:2964–2972
Hosmer DW, Lemeshow S (1980) A goodness-of-fit test for the multiple logistic regression model. Comm Stat A10:1043–1069
Pencina MJ, D’Agostino RB (2004) Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat Med 23:2109–2123
Pencina MJ, D’Agostino RB Sr, Steyerberg EW (2011) Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med 30:11–21
Pencina MJ, D’Agostino RB Sr, Demler OV (2012) Novel metrics for evaluating improvement in discrimination: net reclassification and integrated discrimination improvement for normal variables and nested models. Stat Med 31:101–113
Hosmer DW, Lemeshow S (1995) Confidence interval estimates of an index of quality performance based on logistic regression models. Stat Med 14:2161–2172
Cook NR (2007) Use and misuse of the receiver operating characteristic curve in risk prediction. Circulation 115:928–935
van Hoek M, Dehghan A, Witteman JC et al (2008) Predicting type 2 diabetes based on polymorphisms from genome-wide association studies: a population-based study. Diabetes 57:3122–3128
Talmud PJ, Hingorani AD, Cooper JA et al (2010) Utility of genetic and non-genetic risk factors in prediction of type 2 diabetes: Whitehall II prospective cohort study. BMJ 340:b4838
Vaxillaire M, Veslot J, Dina C et al (2008) Impact of common type 2 diabetes risk polymorphisms in the DESIR prospective study. Diabetes 57:244–254
Balkau B, Lange C, Fezeu L et al (2008) Predicting diabetes: clinical, biological, and genetic approaches: data from the Epidemiological Study on the Insulin Resistance Syndrome (DESIR). Diabetes Care 31:2056–2061
Vassy JL, Mahapatra PD, Meigs JB, et al (2012) Genotype predicts type 2 diabetes in adulthood in a biracial adolescent population. In: American Heart Association Epidemiology and Prevention and Nutrition, Physical Activity and Metabolism 2012 Scientific Sessions, San Diego, CA
Cooke JN, Ng MC, Palmer ND et al (2012) Genetic risk assessment of type 2 diabetes-associated polymorphisms in African Americans. Diabetes Care 35:287–292
Palmer ND, Hester JM, An SS et al (2011) Resequencing and analysis of variation in the TCF7L2 gene in African Americans suggests that SNP rs7903146 is the causal diabetes susceptibility variant. Diabetes 60:662–668
Billings LK, Florez JC (2010) The genetics of type 2 diabetes: what have we learned from GWAS? Ann N Y Acad Sci 1212:59–77
National Cholesterol Education Program (2004) Risk assessment tool for estimating 10-year risk of developing hard CHD. Available from http://hp2010.nhlbihin.net/atpiii/calculator.asp?usertype=prof. Accessed 7 July 2011
Hivert MF, Jablonski KA, Perreault L et al (2011) An updated genetic score based on 34 confirmed type 2 diabetes loci is associated with diabetes incidence and regression to normoglycemia in the diabetes prevention program. Diabetes 60:1340–1348
Acknowledgements
The authors gratefully acknowledge the CARDIA study participants and staff for their valuable contributions. This manuscript has been reviewed by CARDIA for scientific content and consistency of data interpretation with previous CARDIA publications.
This work was presented at the ADA meeting in Philadelphia, USA, June 8–12, 2012.
Funding
JLV is supported by National Institutes of Health (NIH) National Research Service Award grant T32 HP12706 from the Health Resources and Services Administration and the NIH Loan Repayment Program (NIDDK). JB Meigs is supported by NIH grants K24 DK080140 and R01 DK078616. CE Lewis is supported by NIH grant P60 DK07626 to the University of Alabama at Birmingham Diabetes Research and Training Center. The CARDIA study is supported by grant 5 R01 HL078972 from the National Heart Lung and Blood Institute (NHLBI) and was partially supported by contracts N01-HC-48047 (University of Alabama at Birmingham), N01-HC-48048 (University of Minnesota), N01-HC-48049 (Northwestern University), N01-HC-48050 (Kaiser Foundation Research Institute) and N01-HC-95095 (University of Alabama at Birmingham) from the NHLBI/NIH.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
JLV made substantial contribution to the conception and design, analysis and interpretation of the data, contributed to discussion, wrote the manuscript and takes responsibility for the contents of the article. NHD, EKK, MRC and LJRT contributed to interpretation of the data and revised the manuscript for important intellectual content. MF contributed to conception and design, data acquisition and interpretation, and revised the manuscript for important intellectual content. CEL and DSS contributed to acquisition and interpretation of the data and revised the manuscript for important intellectual content. JBM made substantial contribution to the conception and design, analysis and interpretation of the data, and revised the manuscript for important intellectual content. All authors approved the final version of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary materials
Below is the link to the electronic supplementary material.
ESM Table 1
(PDF 110 kb)
ESM Table 2
(PDF 86 kb)
ESM Table 3
(PDF 89 kb)
ESM Table 4
(PDF 88 kb)
Rights and permissions
About this article
Cite this article
Vassy, J.L., Durant, N.H., Kabagambe, E.K. et al. A genotype risk score predicts type 2 diabetes from young adulthood: the CARDIA study. Diabetologia 55, 2604–2612 (2012). https://doi.org/10.1007/s00125-012-2637-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-012-2637-7