Abstract
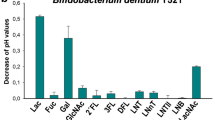
Mucus production is initiated before birth and provides mucin glycans to the infant gut microbiota. Bifidobacteria are the major bacterial group in the feces of vaginally delivered and breast milk-fed infants. Among the bifidobacteria, only Bifidobacterium bifidum is able to degrade mucin and to release monosaccharides which can be used by other gut microbes colonizing the infant gut. Eubacterium hallii is an early occurring commensal that produces butyrate and propionate from fermentation metabolites but that cannot degrade complex oligo- and polysaccharides. We aimed to demonstrate that mucin cross-feeding initiated by B. bifidum enables growth and metabolite formation of E. hallii leading to short-chain fatty acid (SCFA) formation. Growth and metabolite formation of co-cultures of B. bifidum, of Bifidobacterium breve or Bifidobacterium infantis, which use mucin-derived hexoses and fucose, and of E. hallii were determined. Growth of E. hallii in the presence of lactose and mucin monosaccharides was tested. In co-culture fermentations, the presence of B. bifidum enabled growth of the other strains. B. bifidum/B. infantis co-cultures yielded acetate, formate, and lactate while co-cultures of B. bifidum and E. hallii formed acetate, formate, and butyrate. In three-strain co-cultures, B. bifidum, E. hallii, and B. breve or B. infantis produced up to 16 mM acetate, 5 mM formate, and 4 mM butyrate. The formation of propionate (approximately 1 mM) indicated cross-feeding on fucose. Lactose, galactose, and GlcNAc were identified as substrates of E. hallii. This study shows that trophic interactions of bifidobacteria and E. hallii lead to the formation of acetate, butyrate, propionate, and formate, potentially contributing to intestinal SCFA formation with potential benefits for the host and for microbial colonization of the infant gut. The ratios of SCFA formed differed depending on the microbial species involved in mucin cross-feeding.



Similar content being viewed by others
References
Brockhausen I, Schachter H, Stanely P (2009) O-GalNAc glycans. In: Varki A, Cummings RD, Esko JD, et al. (eds) Essentials of glycobiology, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor,
Pelaseyed T, Bergström JH, Gustafsson JK, et al. (2014) The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system Immunol Rev 260:8–20
Tailford LE, Crost EH, Kavanaugh D, Juge N (2015) Mucin glycan foraging in the human gut microbiome Front Genet 6:81
Koropatkin NM, Cameron EA, Martens EC (2014) How glycan metabolism shapes the human gut microbiota Nat Rev Microbiol 10:323–335
Avershina E, Storrø O, Øien T, et al. (2013) Bifidobacterial succession and correlation networks in a large unselected cohort of mothers and their children Appl Environ Microbiol 79:494–507
Sun Z, Zhang W, Guo C, et al. (2015) Comparative genomic analysis of 45 type strains of the genus Bifidobacterium: a snapshot of its genetic diversity and evolution PLoS One 10:e0117912
Bottacini F, Ventura M, Van Sinderen D, Motherway MOC (2014) Diversity, ecology and intestinal function of bifidobacteria Microb Cell Factories 13:1024
Sela DA, Chapman J, Adeuya A, et al. (2008) The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome Proc Natl Acad Sci U S A. 105:18964–18969
Rockova S, Rada V, Nevoral J, Marsik P, Vlkova E, Bunesova V (2012) Inter-species differences in the growth of bifidobacteria cultured on human milk oligosaccharides Folia Microbiol 57:321–324
LoCascio RG, Desai P, Sela DA, Weimer B, Mills DA (2010) Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization Appl Environ Microbiol 76:7373–7381
Turroni F, Duranti S, Bottacini F, Guglielmetti S, Van Sinderen D, Ventura M (2014) Bifidobacterium bifidum as an example of a specialized human gut commensal Front Microbiol 5:437
Milani C, Luigli GA, Duranti S, et al. (2015) Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut Sci Rep 5:15782
Duranti S, Milani C, Lugli GA, et al. (2015) Insights from genomes of representatives of the human gut commensal Bifidobacterium bifidum Environ Microbiol 17:2515–2531
Ruas-Madiedo P, Gueimonde M, Fernández-García M, de los Reyes-Gavilán CG, Margolles A (2008) Mucin degradation by Bifidobacterium strains isolated from the human intestinal microbiota Appl Environ Microbiol 74:1936–1940
Garrido D, Ruiz-Moyano S, Lemy DG, Sela DA, German JB, Mills DA (2015) Comparative genomics reveals key differences in response to milk oligosaccharides of infant gut-associated bifidobacteria Sci Rep 4:5
Egan M, Motherway MOC, Kilcoyne M, et al. (2014) Cross-feeding by Bifidobacterium breve UCC2003 during co-cultivation with Bifidobacterium bifidum PRL2010 in a mucin-based medium BMC Microbiol 14:282
de Vries W, Stouthamer AH (1967) Pathway of glucose fermentation in relation to the taxonomy of bifidobacteria J Bacteriol 93:574–576
Palframan RJ, Gibson GR, Rastall RA (2003) Carbohydrate preferences of Bifidobacterium species isolated from the human gut Curr Issues Intes Microbiol 4:71–75
Schwab C, Ruscheweyh HJ, Bunesova V, Pham VT, Beerenwinkel N, Lacroix C (2017) Trophic interactions of infant bifidobacteria and Eubacterium hallii during L-fucose and fucosyllactose degradation Front Microbiol 8:95
Bunesova V, Lacroix C, Schwab C (2016) Fucosyllactose and L-fucose utilization of infant Bifidobacterium longum and Bifidobacterium kashiwanohense BMC Microbiol 16:248
Reichardt N, Duncan SH, Young P, et al. (2014) Phylogenetic distribution of three pathways for propionate production within the human gut microbiota ISME J 8:1323–1335
Engels C, Ruscheweyh H-J, Beerenwinkel N, Lacroix C, Schwab C (2016) The common gut microbe Eubacterium hallii also contributes to intestinal propionate formation Front Microbiol 7:713
Scott KP, Martin JC, Duncan SH, Flint HJ (2013) Prebiotic simulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro FEMS Microbiol Ecol 87:30–40
Wang JMW, de Souza R, Kendall CWC, Emam A, Jenkins DJA (2006) Colonic health: fermentation and short chain fatty acids J Clin Gastroenterol 40:235–243
Vazquez-Gutierrez P, Lacroix C, Jaeggi T, Zeder C, Zimmerman MB, Chassard C (2015) Bifidobacteria strains isolated from stools of iron deficient infants can efficiently sequester iron BMC Microbiol 15:3
Duncan SH, Hold GL, Barcenilla A, Stewart CS, Flint HJ (2002) Lactate-utilizing bacteria, isolated from human feces that produce butyrate as a major fermentation product Appl Environ Microbiol 70:5810–5817
Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P (2009) Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov Int J Syst Evol Microbiol 52:2141–2146
Matsuki T, Watanabe K, Fujimoto J, et al. (2004) Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria Appl Environ Microbiol 70:167–173
Liu S, Ren F, Zhao L, et al. (2015) Starch and starch hydrolysates are favorable carbon sources for bifidobacteria in the human gut BMC Microbiol 15:54
Vanderhaeghen S, Lacroix C, Schwab C (2015) Methanogen communities in stools of humans of different age and health status and co-occurrence with bacteria FEMS Microbiol Lett 362:fnv092
Stoddard SF, Smith BJ, Hein R, Roller BRK, Schmidt TM (2015) rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development Nucleic Acids Res 43:D593–D598
Větrovský T, Baldrian P (2013) The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses PLoS One 8:e57923
Aziz RK, Bartels D, Best AA, et al. (2008) The RAST Server: rapid annotations using subsystems technology BMC Genomics 9:75
Holden HM, Rayment I, Thoden JB (2003) Structure and function of enzymes of the Leloir pathway for galactose metabolism J Biol Chem 278:43885–43888
Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM (2004) Diversity of microbial sialic acid metabolism Microbiol Mol Biol Rev 68:132–153
Turroni F, Bottacini F, Foroni E, et al. (2010) Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging Proc Natl Acad Sci U S A 107:19514–19519
Turroni F, Milani C, van Sinderen D, Ventura M (2011) Genetic strategies for mucin metabolism in Bifidobacterium bifidum PRL2010: an example of possible human-microbe co-evolution Gut Microbes 2:183–189
Pham VT, Lacroix C, Braegger CP, Chassard C (2016) Early colonization of functional groups of microbes in the infant gut Environ Microbiol 18:2246–2258
Duncan SH, Barcenilla A, Stewart CS, Pryde SE, Flint HJ (2002) Acetate utilization and butyryl coenzyme A (CoA):acetate CoA transferase in butyrate-producing bacteria from the human large intestine Appl Environ Microbiol 68:5186–5190
Naseem S, Konopka JB (2015) N-acetylglucosamine regulates virulence properties in microbial pathogens PLoS Pathog 11:e1004947
Magnusdottir S, Heinken A, Kutt L, et al. (2017) Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota Nat Biotechnol 35:81–89
Schwab C, Gänzle MG (2011) Lactic acid bacteria fermentation of human milk oligosaccharide components, human milk oligosaccharides and galactooligosaccharides FEMS Microbiol Lett 315:141–148
Belenguer A, Duncan SH, Graham Calder A, et al. (2006) Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut Appl Environ Microbiol 72:3593–3599
Moens F, Verce M, de Vuyst L (2017) Lactate- and acetate-based cross-feeding interactions between selected strains of lactobacilli, bifidobacteria and colon bacteria in the presence of inulin-type fructans Int J Food Microbiol 241:225–236
Louis P, Flint HJ (2016) Formation of propionate and butyrate by the human colonic microbiota Environ Microbiol 19:29–41
Midtvedt AC, Carlstedt-Duke B, Midtvedt T (1994) Establishment of a mucin-degrading intestinal microflora during the first two years of human life J Ped Gastroenterol Nutr 18:321–326
Fuchs G (1986) CO2 fixation in acetogenic bacteria: variations on a theme FEMS Microbiol Rev 39:181–213
Liu Y, Whitman WB (2008) Metabolic, phylogenetic and ecological diversity of methanogenic archaea Ann N Y Acad Sci 1125:171–189
Yatsunenko T, Rey FE, Manary MJ, et al. (2012) Human gut microbiome viewed across age and geography Nature 486:222–228
Rey FE, Faith JJ, Bain J, et al. (2010) Dissecting the in vivo metabolic potential of two human gut acetogens J Biol Chem 285:22082–22090
Acknowledgments
Vera Bunesova was supported by SCIEX grant 13.151. The authors thank Alfonso Die for technical assistance and Glycom A/S, Denmark, for supplying NANA.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
ESM 1
(DOCX 59.6 kb)
Rights and permissions
About this article
Cite this article
Bunesova, V., Lacroix, C. & Schwab, C. Mucin Cross-Feeding of Infant Bifidobacteria and Eubacterium hallii . Microb Ecol 75, 228–238 (2018). https://doi.org/10.1007/s00248-017-1037-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-017-1037-4