Abstract
Despite the continuous evolution of positron emission tomography (PET) technology, the spatial resolution (SR) of PET images remains poor (4–6 mm) compared with that of computed tomography (CT) and magnetic resonance imaging (<1 mm). The reasons for this lie intrinsically in the physics behind PET and in the scanner design. This poor SR and the consequent partial volume effect (PVE), affecting image quality and the estimation of radioactivity concentration in small structures, complicate the anatomical localisation of focal radiotracer uptake signals and the application of a quantitative approach in the evaluation of PET studies. Methods for PVE compensation are therefore needed to improve the quality and quantitative accuracy of PET images. The simplest of the PVE compensation methods is the one based on recovery coefficients. Other techniques exploit high-resolution structural information extracted from co-registered CT or MR images to correct for the PVE. Others still are aimed instead at recovering the degradation effect by exploiting the point spread function of the PET system within the reconstruction process or using post-processing deconvolution algorithms. Regardless of the method employed, PVE compensation techniques generally assume motion-free PET data. In neurological studies, this assumption may be verified; in cardiological and oncological applications, on the other hand, organ/lesion motion and the PVE should necessarily be taken into account and corrected for sequentially or simultaneously. This paper deals with the PVE, its impact on qualitative and quantitative PET imaging, and possible approaches for PVE compensation.
Similar content being viewed by others
Introduction
Molecular imaging by positron emission tomography (PET) is currently widely used for the detection and functional characterisation of several pathologies. It allows in vivo non-invasive assessment of areas with increased or decreased functionality (e.g. blood flow, metabolism, receptor density etc.) [1]. One particular strength of the PET technique is its high sensitivity, which allows the detection of small amounts of radiotracer, even at picomolecular level. Conversely, spatial resolution (SR) is its weakest point. Despite the continuous technological evolution of PET system designs, SR remains limited to about 4–6 mm [2].
A consequence of this finite SR is the so-called partial volume effect (PVE), in which PET images are affected, both qualitatively and quantitatively, by mixing of the radioactivity coming from neighbouring regions. Blurring due to SR reduces the image contrast, thus limiting the detectability of small lesions and preventing precise anatomical localisation of focal radiotracer uptake. The development and clinical diffusion of hybrid PET/computed tomography (PET/CT) and more recently PET/magnetic resonance imaging (PET/MRI) systems have provided ways of overcoming this lack of anatomical detail [3, 4]. From a quantitative point of view, the poor SR results in a biased estimation of regional radioactivity concentration. As measurement of regional radiotracer uptake has been proven to be useful in differentiating normal and pathological patterns [5], in assessing response to therapy [6], and in predicting disease aggressiveness [7], compensation for PVE is fundamental.
Many PVE correction techniques have been proposed in the literature, with the aim of improving PET image quality and quantitative accuracy [8–12]. PVE correction techniques, grouped into different categories, have been described in several reviews and topical papers comparing, for example, reconstruction-based vs. post-reconstruction or image-based techniques [9, 10] and region-based (RB) vs. voxel-based (VB) techniques [8].
This paper adopts the classification into RB and VB techniques. RB methods are those aiming to achieve a more accurate quantification of the radioactivity concentration in a target. They are applied to specific structures of interest and do not generate PVE-corrected images. VB techniques, on the other hand, seek to compensate for PVE through recovery of the system’s SR at voxel level (generally over the whole image) and aim to generate PVE-recovered images. This classification into RB and VB techniques was adopted with a view to assessing the impact of PVE correction on different patterns of radioactivity distribution, local (e.g. oncology) or more widely distributed (e.g. neurology).
The simplest of the PVE correction methods is the one based on recovery coefficients. Other techniques use high-resolution structural information extracted from co-registered CT or MR images to correct for PVE. Others still aim to recover the degradation effect by exploiting the point spread function of the PET system within the reconstruction process or through the use of post-processing deconvolution algorithms. Sophisticated state-of-the-art PVE compensation methods exploit PET/CT and PET/MRI hybrid scanners, 4D gating tools, flexible iterative reconstruction algorithms and high-performance computing systems. The diffusion of PVE compensation methods in the clinical setting demands extensive validation, optimisation for specific clinical applications, and embedding within clinical image processing software packages.
This paper deals with PVE, its impact on qualitative and quantitative PET imaging, and possible PVE compensation approaches.
PET spatial resolution and the point spread function of the pet system
The SR of a PET imaging system is characterised by its point spread function (PSF), i.e. the response of the imaging system to a radioactive point source. The full width at half maximum (FWHM) of a profile drawn through the PSF is generally taken as a measurement of SR [13, 14]. The PSF accounts for the PET signal degradation due to the emission and detection of the radiation and to the processing of recorded data. As some of the degradation effects are spatially variant, the FWHM of the PSF increases from the centre towards the edges of the scanner field of view (FOV) [14] (Fig. 1).
Typical PET PSF images obtained at different radial positions (0, 5, 10, 15, 20 cm of axis) within the PET FOV. The response of the system is variant, typically narrowest in the centre of the scanner, increasing towards the edges of the FOV. Two profiles drawn through the radial direction of the PSF: on axis (left) and 20 cm off axis (right)
Three-dimensional (3D) asymmetric Gaussian functions, characterised by different FWHMs in radial, tangential and axial directions, are commonly used to describe the spatial variation of the PSF in the PET FOV.
In some conditions, when the PSF variation can be considered small compared with the variation over the whole FOV, a spatially invariant PSF represented by a 3D symmetric Gaussian function can be used.
To obtain an accurate estimation and accurate modelling of the PSF, several techniques have been developed, like Monte Carlo (MC) simulations, analytical models, and experimental measures [13]. MC simulations have been extensively used to assess the detector response and to optimise the design of PET systems. In fact, the MC technique allows simulation of the physical processes involved in radiation emission and interaction with matter, and can thus take into account for (1) positron range, (2) non-collinearity of gamma rays, and (3) detector response in terms of inter-crystal penetration and inter-crystal scattering [13]. The main limitations of MC methods are the long computation time and the need for a detailed knowledge of the system design. Analytical methods also require knowledge of the system design, but, being faster than MC simulations, they represent a valid alternative solution for the assessment of PET detector response. Most of the analytical methods describe inter-crystal penetration as the primary interaction of the radiation with the detector. More complete analytical models also account for the inter-crystal scattering effect [13]. Positron range and gamma ray non-collinearity can also be described analytically, but are most commonly accounted for as convolution functions. Experimental measurements are conceptually the easiest way to obtain the PSF of a PET system. However, in practice, obtaining precise and accurate point source measurements is not an easy task. Indeed, multiple (theoretically adimensional) “identical” point sources with sufficiently high activity concentration are needed to measure PSF variation in the scanner FOV in a reasonable time. Furthermore, the point sources must be precisely positioned within the scanner FOV. A smart and efficient solution has been proposed by Kotasidis et al. [15], who produced reproducible radioactive point sources using a standard printer. Panin et al. [16] instead implemented a robotised system which can perform thousands of measurements in an automatic micro-controlled way.
To obtain high statistics measures without exposing operators to high radiation doses, low activity long-lived isotopes (e.g. 68Ge, 22Na) are commonly used [16, 17]. However, this solution, while practical, results in non-optimal PSF models, as the positron range does not exactly correspond to that of the isotopes used in clinical studies (18F, 11C, 13N, 15O, 82Rb). This aspect is particularly important for high-resolution PET tomographs and for preclinical PET systems. Kotasidis et al. [18] recently proposed a practical strategy for obtaining isotope-specific PSFs. The printing technique used by these authors was adopted to simultaneously acquire multiple point sources of 18F and 11C.
The partial volume effect (PVE)
The effect produced by the finite SR can be mathematically expressed, in the image space, as a 3D convolution between the true spatial distribution of radioactivity and the PSF of the PET system. The blurring effect of the PSF in fact spreads the contribution of each ideal point source of radioactivity over a wider volume. The content of each pixel is thus the sum of the contributions from neighbouring blurred point sources. This mix of signals is called the partial volume effects (PVE) [19–21]. The PVE becomes particularly important when the target of interest is smaller than 2–3 times the system’s FWHM, as it is in almost all brain structures, the myocardial wall, and small tumours.
With respect to the PVE, four clinical conditions must be considered: (1) hot target without a surrounding active background (Fig. 2a), (2) hot target surrounded by a background with lower activity, (3) hot target surrounded by a higher active background, and (4) cold target surrounded by an active background (Fig. 2b). In the first case (1) the PVE results in an underestimation of the radioactivity concentration of the hot target and overestimation of its volume, as a result of the spreading from the target region into the background (spill-out effect). In the second case (2), a spread from the background into the region of the target is also present (spill-in effect). In the last two cases (3, 4), a spill-in effect dominate, resulting in overestimation of the target radioactivity concentration and underestimation of its volume.
a Simulation of the finite spatial resolution of an imaging system. Top row 2D circular (binary) objects with diameters equal to (from left to right): 1, 2, 3 and 4 times the FWHM (5 mm) of the imaging system. Central row the same circular objects convolved with the PSF of the imaging system showing (qualitatively) the blurring effect due to the finite spatial resolution. Bottom row profiles drawn through the binary (broken lines) and the convolved (continuos line) objects. b Simulation of the finite spatial resolution of an imaging system for a circular object (CO diameter = 5 mm) in a circular uniform background (UB diameter = 30 mm). Top row (left) CO intensity = 100, UB intensity = 50; (centre) CO intensity = 50, UB intensity = 100); (right) CO intensity = 0, UB intensity = 100). Central row the same circular objects convolved with the PSF of the imaging system (FWHM = 5 mm) showing the blurring effect due to the finite spatial resolution. Bottom row profiles drawn through the original (broken lines) and the convolved (continuos line) objects
The combined effect of spill-out and spill-in is referred as spill-over, and represents the “total” cross-contamination between adjacent structures with different radiotracer uptake.
Another PVE-associated effect affecting the accuracy of signal quantification is due to the finite dimensions of the image voxel. The larger the voxel size, the higher the probability of it including two or more different types of tissue, whose contributions are averaged in a confounding signal. This effect is called tissue fraction (TF) [8]. To reduce the TF effect, small voxels should be used, which, however, increase the statistical noise. Voxel size therefore needs to be carefully chosen to balance TF and noise level.
Recovery of spatial resolution and PVE correction techniques
Region-based methods
PVE correction by recovery coefficients
Of the various PVE correction methods, the simplest and most practical to implement in a clinical setting are those based on multiplicative numerical factors (recovery coefficients, RCs). RCs can be derived from PET experimental measurements of radioactive objects simulating the anatomical structures under investigation (e.g. small tumour lesions). These are generally spherical objects of known diameter, filled with a known radioactivity concentration that are then placed in a cold background [19–21]. The RC is the ratio between PET-measured radioactivity and actual radioactivity concentration within the spheres.
This simple approach was applied for the PVE correction of 18F-fluorodeoxyglucose (18F-FDG) PET images of oncological lesions in real patients, since radioactive spheres were considered suitable for simulating metabolic active oncological lesions [22]. Unfortunately, given the experimental set-up used to calculate the RC, the method was able to compensate only for the spill-out effect from the lesions into the surrounding background of the patient’s body, not accounting for the spill-in effect from the background into the lesion. More realistic models considering a hot background surrounding the target were then implemented; these involved measuring and combining RCs derived from hot objects in cold backgrounds, cold objects in hot backgrounds, and hot objects in hot backgrounds. In this way, both spill-out and spill-in effects could be accounted for. In fact, even when considering a set of curves reporting experimentally measured RCs as a function of lesion size and for different target-to-background radioactivity concentration ratios (T/B), to apply an RC-based PVE correction method to real clinical studies, one would still need to know the actual T/B and the actual volume of the target region, so as to allow the correct RC value to be properly sampled from the RC curves. Unfortunately, the PVE makes it impossible to measure either the lesion volume or the T/B ratio from PET clinical images. Srinivas et al. [23] implemented an RC-based PVE correction method specifically for oncological studies, i.e. measuring RCs from hot spheres in hot background experiments. RC curves were thus obtained for measured T/B (T/Bm), but unfortunately as a function of the actual sphere volume. As Srinivas et al. [23] suggested, lesion volume can be measured on registered anatomical CT or MR images, but unfortunately PET lesions are not always visible on CT images. Furthermore, the metabolic PET volume may be different from the CT/MRI anatomical volume.
Another issue regarding RC-based PVE correction methods is the need to use the same technique to measure radioactivity concentration both in the set-up of the method (generation of the RC curves) and in the estimation of the lesion radiotracer uptake. An operator-dependent technique, such as drawing a region of interest (ROI) around the lesion, can introduce inter-observer variability. On the other hand, an operator-independent technique, such as considering the maximum pixel value in the lesion, is more sensitive to the noise level of PET images, and thus requires optimisation strategies and accurate validation [8].
Gallivanone et al. [24, 25] proposed an RC-based PVE correction technique, attempting to address these critical issues. Their method is based on experimental studies of hot spheres in hot backgrounds at different T/B ratios and on the generation of RC curves as a function of T/Bm and measured sphere volume (Fig. 3). Given an oncological lesion, T/Bm and lesion volume are measured and the proper RC is sampled from RC curves.
Recovery coefficient (RC) curves as function of tumour-to-background ratio (T/B) and tumour sphere-equivalent diameter (d) measured on PET images. Courtesy of Gallivanone et al. [24]
PVE correction from PET raw data
A different approach to RB PVE correction consists of working in the sinogram rather than in the image domain. This approach was originally proposed by Huesman et al. [26] for the quantification of PET dynamic studies. With this method, a ROI defined on the structure of interest, is forward projected in the sinogram space, convolved with the filter function used for the image reconstruction and then back projected. The advantages of the method are the low computation burden, as reconstruction of the whole dynamic dataset is not required, and the possibility of easily computing the statistical uncertainty of the radioactivity quantification in the ROI. The Huesman technique was improved by Muzic et al. [27], who used a 2D inverse filtering to compensate for the SR degradation.
Carson [28] proposed a maximum likelihood expectation maximisation method for ROI quantification in the sinogram domain, taking into account the Poisson statistics of noise and fully compensating for PVE, by modelling the physical factors contributing to the projection measurements. An alternative approach which uses a least squares (LS) approach was proposed by Formiconi [29] and assessed by Vanzi et al. [30] on phantom and clinical studies of the dopaminergic system. The LS method proved more accurate than the conventional strategy of measuring ROIs on the reconstructed images. Using a dual modality system, Moore et al. [31] proposed a technique allowing full PVE compensation within a volume of interest (VOI) surrounding a target lesion. This method requires segmentation of co-registered anatomical images (CT or MR) into a few (typically 2–4) tissue types within the defined VOI. The activity concentration within each segmented tissue is then iteratively estimated from PET projection data by maximising a likelihood function, taking into account the statistical noise and the scanner SR.
PVE compensation using the geometric transfer matrix method
The geometric transfer matrix (GTM) method, proposed by Rousset et al. [32], is a RB PVE correction algorithm that estimates the average uptake in multiple ROIs defined on spatially co-registered anatomical images. The technique requires segmentation of the anatomical image into a set of N non-overlapping ROIs, each assumed to contain uniform PET activity. Each ROI, defined as a binary map, is then blurred with the system PSF to obtain the corresponding regional spread fraction (RSF). Once the fractional contribution (ω i,j ) of each RSF j into each ROI i has been computed, a system of N linear equations relating PET mean values (M PET) in ROIs to the unknown true PET activities (T PET) can be defined as:
The solution of the system of linear equations can thus be obtained as:
The results of the GTM method are the true average activity concentrations in each ROI, corrected for the spill-over between adjacent regions.
Du et al. [33] proposed a variation of the GTM method, named perturbed GTM (pGTM), which, in the computation of the \( \omega_{i,j} \) elements, accounts for the non-linear effects due to the iterative reconstruction, especially when a resolution recovery model (PSF) is included in the reconstruction scheme. The pGTM method was proven to reduce the bias in the measured activity compared to GTM on both MC simulations and experimental data.
Recently, a further variation of the GTM method was proposed by Sattarivand et al. [34]. Like GTM, their technique, named symmetric GTM (sGTM), defines a system of N linear equations (one for each ROI) but the matrix of the coefficients (\( \omega_{i,j} \)) contains the fractional contributions of RSF j into RSF i , instead of into ROI i . When compared with GTM, the sGTM method showed: (1) similar accuracy, (2) better precision for small objects, (3) inferior noise propagation, and (4) better robustness vs. mis-registration errors or errors in the PSF estimate.
Voxel-based methods
Recovery of spatial resolution and PVE correction by image reconstruction
A direct method to compensate for PVE is to recover the SR of the PET system within the image reconstruction process. Iterative reconstruction algorithms are particularly suitable for this purpose as they allow physical models describing the emission and detection of radiation, as well as design characteristics of the PET system, to be included in the reconstruction scheme [35]. Furthermore, statistical iterative algorithms can also account for the Poissonian nature of the noise associated with the measured data [36].
The modelling of radiation emission/detection and the handling of statistical noise allow a significant qualitative and quantitative enhancement of the reconstruction. From this perspective, inclusion of the PSF in the reconstruction algorithm should allow recovery of the SR and, thus compensation for the PVE (at least the spill-over effects) [16, 17, 38].
Unfortunately, to obtain a significant recovery of the degraded PET signal, iterative algorithms should be run up to convergence, and thus for a high number of iterations. However, a high number of iterations have two drawbacks: (1) a long computation time, impractical in the clinical setting; (2) an increase of the image noise. Stopping rules and smoothing filters are therefore commonly used, which, however, reduce the amount of recovery and induce non-uniform resolution across the FOV [36]. Furthermore, as iterative algorithms are not linear, the level of SR recovery is dependent on the specific radioactivity distribution. Another recognised effect of PSF-based reconstructions is an over-enhancement of sharp transitions (edges) in the reconstructed images [13, 39].
Notwithstanding these limitations, reconstruction algorithms accounting for the PSF have been shown to lead to improved quality and the quantitative accuracy, both with simulated and with experimental PET data [16, 17, 38]. Today, they are available on PET/CT scanners and increasingly used across all clinical applications [40–44].
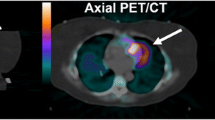
Following the recent introduction of time-of-flight technology (TOF), TOF information can also be incorporated into the reconstruction algorithm. TOF makes it possible to reduce image noise, in particular in large patients, and accelerate convergence. The combination of TOF and PSF thus represents a further potential step towards improvement of PET reconstruction [45–49] (Fig. 4).
18F-FDG PET/CT. Top CT image showing a small nodule in the lung. Bottom two PET images reconstructed using a 3D-OSEM algorithm without (left) and with (right) TOF + PSF information. Note the improved resolution and contrast as well as the quantitative change in the SUVmax value when TOF + PSF information is accounted for in the reconstruction algorithm
Furthermore, the anatomical information contained in high-resolution CT or MR images can support and constrain PET image reconstruction to obtain better SR recovery and better noise control [37]. Spatially co-registered anatomical images can in fact provide relevant information about the tissues underlying the functional signal: (1) borders of regions with homogeneous anatomical characteristics, (2) transitions between regions with different anatomical characteristics. In the first case, the noise reduction can be tuned to smooth the PET signal within the region boundaries; in the second, a preservation of the PET signal can be imposed in the regions of transition between different types of tissues [37].
Several methods supporting the use of anatomical information in PET reconstruction have been proposed over the years, opening the way for significant improvements in image quality and quantitative accuracy. However, to our knowledge, there has not yet been an extensive clinical evaluation of such algorithms to support their use in clinical settings.
Recovery of spatial resolution and PVE correction by image deconvolution
Another approach to SR recovery, similar to image reconstruction, but simpler to implement, is image deconvolution [50]. Image deconvolution is a post-reconstruction restoration technique that aims to recover SR degradation through knowledge of the system’s PSF. An observed (degraded) image \( o(\bar{x}) \) can in fact be mathematically described as the convolution of the true image \( i(\bar{x}) \) with the PSF of the imaging system \( h(\bar{x}) \):
where \( \bar{x} \) is a point of coordinate \( (x,y,z) \), \( \otimes \) is the convolution operator, and \( h(\bar{x}) \) is assumed as shift invariant. However, because of the noise component, which can be accounted for as an additive term in the previous equation, the deconvolution problem is ill-posed, i.e. a unique stable solution does not exist. Iterative techniques are therefore commonly used. The most popular ones are: (1) the Van Cittert (VC) algorithm, and one of its variations, the re-blurred Van Cittert (R-VC), and (2) the Richardson–Lucy (RL) algorithm [50].
The VC algorithm belongs to the family of constrained iterative algorithms, whose goal is to obtain, through an iterative process and under a positive constraint, a solution that, convolved with the PSF, generates the observed image. The algorithm assumes that the noise in the image is negligible and seeks a solution that minimises the LS criterion:
The solution is expressed by the following equation:
where \( \overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\frown}$}}{o}_{k} (\bar{x}) \) and \( \overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\frown}$}}{o}_{k + 1} (\bar{x}) \) are the solutions at k and k + 1 iterations, and α (α = [0, 2]) is a parameter controlling the convergence rate. Unfortunately, a common problem of deconvolution methods is that of noise amplification as the iterations proceed. The R-VC method has been proposed to mitigate this effect, using the following updating scheme:
where \( h( - \bar{x}) \) is the flipped PSF. R-VC performs an extra convolution to stabilise the deconvolution process; in this way it achieves better convergence and lower noise sensitivity [50].
The RL method instead belongs to the family of statistical deconvolution methods, which exploit knowledge of the random nature of noise in the solution search. The RL algorithm assumes that the image is originated from a Poisson process and attempts to iteratively maximise the data likelihood by means of the expectation maximisation algorithm. The formulation of the RL algorithm is:
where the division and multiplication are element-wise.
The RL algorithm (Fig. 5) is similar to the VC one. The difference lies in the updating term: in VC, the update is additive and the updating term depends on the difference between the observed image and the convolution of the current solution with the PSF; in RL, the update is multiplicative and the updating term depends on the ratio between the two quantities. A further convolution is applied to this ratio before the update.
Example of image deconvolution by Richardson–Lucy (RL). Upper image original PET image reconstructed by conventional 3D-OSEM algorithm. Bottom row RL deconvolved images at 3, 5, 10, 20 iterations. A Gaussian shift invariant PSF was used for the SR restoration. Note the SR recovery achievable, but also the high increase in the noise as the iterations proceed
Within the context of PVE correction, deconvolution techniques have recently been used by several groups to compensate for the spill-over effects (spill-in and spill-out) [51–55].
Recovery of spatial resolution and PVE correction by multi-resolution approach
A different method of generating PVE-corrected images was proposed by Boussion et al. [56]. The method is a post-reconstruction enhancement technique which, for the PVE correction, uses information extracted from spatially co-registered high-resolution anatomical images. The method assumes, as mandatory, that a positive correlation exists between the intensity values of functional PET and anatomical images over the whole volume. By means of wavelet transform and multi-resolution analysis, high spatial frequencies are transferred from anatomical to functional images, thus recovering the SR. The wavelet transform, in fact, allows a joint representation of the spatial and frequency domains in the same image. This kind of representation can be obtained at different levels (layers) of SR. Therefore, once the common level of resolution (in which both wavelet images have the same PSF-FWHM) has been established, the details from the high-resolution image are extracted (as wavelet coefficients), transformed (according to a defined linear model) and then integrated (added pixel-to-pixel) into the low-resolution PET image set. PSF is assumed to be shift invariant and identical in all images. The method has been shown to be able not only to improve the PET image, but also to quantitatively adjust the intensity of PET radioactivity concentration, especially at organ edges and for small sources. However, artefacts arise in regions where the correlation between anatomical and functional details is lacking.
The method has recently been improved by Le Pogam et al. [57], who extended the original implementation from 2D to 3D, thus taking into account the intrinsic 3D nature of the PVE. Furthermore, the original requirement of total correlation between functional and anatomical information was relaxed in favour of a local application of the method, wherein the model parameters can be adapted to the specific regional characteristics.
The coupling between wavelet decomposition and multi-resolution analysis of functional and structural information has also been proposed by Shidahara et al. [58] on 18F-FDG and 11C-raclopride brain studies. The technique, named “structural and functional synergy resolution recovery”, uses an anatomical probabilistic brain atlas as the source for the high-resolution structural information to be integrated into PET images. The subdivision of the brain atlas into anatomical districts allows a local analysis and thus relaxes the need for functional/structural correlation (Fig. 6).
PVE correction by partition methods using anatomical images
Partition methods are post-processing PVE compensation techniques, whose common characteristic is that they require the segmentation of spatially co-registered anatomical images into two or more tissue types. PVE correction is then applied on a voxel basis within each segmented region/tissue.
One of the first partition methods was proposed by Videen et al. [59] to correct for the PVE caused by cerebral atrophy (PVE-atrophy). With this method, the anatomical image has to be segmented into two classes: brain and non-brain. The brain mask is then convolved with the scanner PSF to create a brain tissue image of RCs. The pixel-by-pixel ratio between the original (degraded) PET image and the image of the RCs produces the PVE atrophy-corrected PET image. Videen’s work, originally implemented in 2D, was subsequently extended in 3D by Meltzer et al. [60] (Fig. 7). An improvement of the Videen/Meltzer method was proposed by Müller-Gärtner et al. [61] (MG) to compensate for the PVE between brain structures with high and low activity (e.g. grey and white matter) (Fig. 7). The MG method requires segmentation of MR images into three labels: grey matter (GM: X grey), white matter (WM: X white) and cerebrospinal fluid (CSF: X csf). The method assumes that the radioactivity concentration in WM and CSF is constant and known; the mean values (\( \bar{I}_{\text{white}} \), \( \bar{I}_{\text{csf}} \)) thus have to be measured in regions of the reconstructed PET images large enough not to be affected by the PVE. The radioactivity concentration in the GM, \( \bar{I}_{\text{grey}} \) is estimated as follows:
where \( I_{\text{obs}} \) is the radioactivity concentration in the observed PET image and \( h \) is the system’s PSF.
Example of PVE correction techniques by different partition methods: Top row CT image of the Hoffman brain phantom (left) and the corresponding PET images obtained with a GE Advance PET tomograph. Bottom row PVE-corrected images using: Meltzer, Müller-Gärtner and Alfano method. As can be seen the radioactivity distribution in the grey matter, well defined and uniform in the CT image, does not seem to be homogeneous in the (degraded) PET image due to the PVE. In the PVE-corrected images, depending on the method used, such homogeneity of the radioactivity distribution is much better recovered both qualitatively and quantitatively. Courtesy of Harri MA, MSc, Turku PET centre (color figure online)
The GM PVE-corrected image is therefore obtained by subtracting, from the observed (degraded) PET image, the spill-in contributions of WM and CSF, and then by correcting for the GM spill-out.
The Meltzer and MG methods have been used in several studies on aging to account for the potential confounding effect due to the presence of atrophy on the measurements of cerebral blood flow (CBF) [62] and glucose metabolism [63, 64]. In fact, previous PET studies, not taking into account the PVE due to atrophy, inconsistently reported reductions in CBF and glucose metabolism with aging. PVE correction removed such negative correlations, thus indicating that in healthy individuals, CBF and glucose metabolism may not decline with aging [62–64]. A true glucose metabolism reduction after PVE correction was instead observed in Alzheimer’s disease (AD) patients [65].
The MG method corrects for the PVE in GM due to the activity spill-out towards WM and CSF and to the activity spill-in coming from WM and CSF. The PVE between different GM areas is not taken into account. In this sense, the MG method intrinsically assumes an uptake homogeneity in GM and therefore does not allow an accurate PVE compensation in brain structures presenting heterogeneity in GM tissue (e.g. subcortical nuclei). To account for this, Metzler et al. [66] extended the original MG model, by introducing a fourth compartment (a VOI, \( X_{\text{VOI}} \) in the GM, such that \( \bar{I}_{\text{VOI}} \) is different from \( \bar{I}_{\text{grey}} \)). The PVE-corrected \( \bar{I}_{grey} \) value is estimated as in the MG method and then used to calculate the PVE-corrected \( \bar{I}_{\text{VOI}} \) activity as:
An adaptation of the MG method to cardiac studies was recently proposed by Du et al. [67]. Their method requires delineation of the left ventricular (LV) wall and LV blood pool on contrast-enhanced CT images co-registered with the PET images. The whole LV myocardium is therefore treated as a single VOI in which a uniform uptake is assumed. This assumption can introduce errors in the spill-out compensation, when uptake defects are present in the myocardium. To account for this possibility, similarly to Meltzer’s method [66], an additional VOI, representing the defect, can be considered. It is worth noting that, because of the cardiac motion, the PVE correction must be performed on single gated images (e.g. end-diastolic, end-systolic phase).
Taking the MG technique as a reference, other methods have been proposed to improve the weaknesses connected to its assumptions. For example, the MG technique assumes it is measuring a WM value free from PVE and takes this value as the mean WM over the entire brain volume. Rousset et al. [68] proposed a modification of the MG method (modified MG) in which a WM mean uptake corrected for PVE is estimated by means of an initial GTM step [69]. A further improvement, combining VB and RB approaches, was proposed by Alfano et al. [70] with the aim of reducing the noise level on PVE-corrected images (Fig. 7).
The MG method and its extensions are suitable for estimating the PVE-corrected image inside a specific ROI. Yang et al. [71] proposed a method for estimating a PVE-corrected image over multiple ROIs. Their method requires segmentation of the spatially co-registered anatomical images into a defined number of tissues (labels) and a priori knowledge of the activity concentration ratios between different tissues. An image (\( I_{\text{pure}} \)) is thus created by assigning relative activity concentration values to each label. A simulated PET image (\( I_{\text{simulated}} \)) is then generated by convolving \( I_{\text{pure}} \) with the system’s PSF. The PVE-corrected image (\( I_{\text{corrected}} \)) is finally calculated by applying, pixel-by-pixel, the following relation:
where \( I_{\text{measured}} \) is the reconstructed PET image.
Thomas et al. [72] recently proposed an improvement of the Yang method [71], coded as a region-based voxel-wise (RBV) PVE correction, which, instead of assuming a priori knowledge of the tissue activity, estimates the \( I_{\text{simulated}} \) image by means of a GTM approach.
As for RB methods, Erlandsson et al. [73] proposed a partition-based PVE correction technique for application in the projection rather than in the image domain. The method requires that the image can be subdivided into regions, each assumed to have a uniform activity distribution. PVE correction is performed iteratively in combination with a filtered back-projection (FBP) algorithm accounting for the PSF of the detection system. At the nth iteration the algorithm generates a piece-wise constant image corrected for PVE over the set of pre-defined anatomical regions, as follows:
where \( p_{\text{measured}} \) is the PET-measured projection data, FBP{·} represents reconstruction by FBP, A(·) the average operator, F{·} the simple forward-projection, and F AR(·) the forward-projection with non-uniform attenuation and distance-dependent resolution. To obtain better control of noise, the method was recently improved by Erlandsson et al. [74], who substituted FBP with the ordered subsets expectation maximisation (OSEM) algorithm.
Accuracy, precision and robustness of spatial resolution recovery and PVE correction techniques
Each of the considered PVE compensation methods has its own strengths and weaknesses and must therefore be assessed in terms of accuracy, precision and robustness before being used for a specific clinical application (e.g. single study, comparative study, group study). In particular, it is always important to evaluate the trade-off between the bias reduction in the quantitative estimation of the radioactivity concentration and the potential increase in the variability of the results due to the errors associated with the correction procedure. The potential sources of errors that have to be accounted for are: (1) mis-determination of the PSF, (2) noise amplification, (3) mis-determination of reference data (e.g. WM in the MG method), (4) mis-registration between anatomical and functional images, (5) mis-segmentation of anatomical images, and (6) signal heterogeneity. Each of these potential sources of errors may have a different impact on each technique and even on each anatomical district. It is therefore important to assess the sensitivity of each method to each possible source of errors as well as to combinations of these sources (e.g. mis-registration and mis-segmentation) [75, 76].
Partial volume effect correction in the context of kinetic modelling
A peculiarity of PET is that it offers the possibility of quantifying not only the concentration of radioactivity but also the absolute tracer uptake, and therefore makes it possible to describe important physiological processes like blood flow, regional glucose metabolism, receptor density etc. [1, 77, 78]. Quantitative studies aimed at quantifying the uptake rate have traditionally been exploited in the neurological and cardiac research fields. More recently, interest in this approach has also arisen in oncology for better characterisation of tumours [79]. To perform quantitative studies, a dynamic PET scan and a mathematical model describing the radioligand kinetics are needed [77, 78]. The principal components of a kinetic model are the “input function” (IF), representing the amount of tracer in the arterial blood over the time of the study, and the “time-activity curve” (TAC) which is the radioactivity in the tissue of interest measured on the reconstructed images of each dynamic frame. To provide a reliable estimate of the kinetic parameters, both the IF and the TAC should be free from artefacts and not affected by the PVE. The IF can be directly sampled from a peripheral artery of the patient. However, if an artery (e.g. peripheral) of the patient is visible in the acquired FOV, the IF can be measured on the PET image (image-derived input function, ID-IF), thus avoiding the invasiveness of direct sampling. In cardiac studies, the ID-IF is generally measured in the left ventricle, atrium or aorta. However, the accuracy of this measure is limited by the PVE due to scanner SR, heart motion and patient’s respiration. In neurological studies, the ID-IF can be measured if the carotid arteries are visible in the reconstructed FOV. However, the measure is significantly affected by PVE because of the small size of the carotid arteries (~5 mm). The same problem arises in oncological studies in which the ID-IF is measured in visible peripheral arteries. Strategies to obtain ID-IFs which also account for the PVE have been proposed by several groups with good results [80–85]. In particular, one of the first attempts to obtain an ID-IF in brain studies was made by Litton et al. [80] who manually drew ROIs over the internal carotid arteries on spatially co-registered MR images. The ROIs were then transferred on the PET images. PVE correction was performed using RCs, computed assuming the performances of the scanner and a 5 mm diameter of the carotid arteries. Chen et al. [81] derived the IF from carotid arteries directly on 18F-FDG PET dynamic images. In this case, early dynamic PET frames were summed and used to identify carotid arteries where the ROIs could then be drawn. To correct for PVE, the early summed images were superimposed over the last PET frame, where a tissue ROI was drawn around the carotids. PVE correction was performed by solving a linear model where each PET measure was assumed to be a combination of two components: the true radioactivity from the blood and the radioactivity from the surrounding tissues. More recently, Mourik et al. [82] also evaluated the possibility of obtaining an ID-IF directly from dynamic PET data from 11C-flumazenil brain studies. ROIs were defined on blood volume images, derived from the summation of early time frames (from 15 to 45 s), on which the carotid arteries could easily be identified. ID-IF was corrected for the PVE using a PSF-based reconstruction algorithm. Croteau et al. [83], using a hybrid PET/CT system, proposed a method of obtaining the ID-IF from carotid and femoral arteries. The technique requires knowledge of the artery dimensions, which can be obtained from a high-resolution CT image. PVE compensation was then performed by RCs. Hackett et al. [84], also using a PET/CT system, described a method of estimating the IF from reconstructed images of dynamic 18F-FLT PET/CT studies of the head and neck. PVE was also accounted for in the solution of the kinetic model from data obtained using the GTM method with ROIs outlined on CT images and then transferred on PET data. Despite the good results reported in the above-mentioned studies, obtaining an ID-IF remains a very challenging task; therefore, prior to clinical implementation, each method should be carefully tested and its limitations thoroughly evaluated with respect to the specific application [85].
Besides the IF, the TACs of the tissues of interest are also affected by problems due to limited SR. To account for PVE in TAC sampling, two strategies can be followed: (1) one of the previously described PVE techniques can be applied on each frame of the dynamic PET dataset before sampling the TAC; (2) the PVE can be accounted for in the kinetic model, by considering additional free parameters to be fitted together with the physiological parameters [86, 87]. The second strategy has been widely used both in cardiac and brain studies.
Recently, a new approach to PET quantification by kinetic modelling, referred to as 4D PET image reconstruction, was proposed. This new approach is based on a more comprehensive use of the spatiotemporal information contained in the PET dynamic data. In particular, a priori knowledge is exploited by making use of temporal filtering, wavelets, principal component analysis or direct kinetic modelling in the reconstruction [88]. In the currently investigated 4D reconstruction protocols, the system’s PSF is also taken into account.
Organ motion
All the previously described PVE correction techniques implicitly assume immobility of the target both in PET and in corresponding spatially co-registered anatomical images, especially when the anatomical information is used to support the PVE compensation. However, this condition is often lacking, since most of the organs in the body (heart, lungs, kidneys, spleen, stomach, bladder, etc.) are affected by movement, mainly due to patient respiration [89]. The situation is even worse in cardiac studies where cardiac and respiratory motions are both present. In addition, voluntary/involuntary patient movements due to an uncomfortable position or to disease-related pain can occur during the study.
A direct consequence of target motion is further degradation of the SR and consequently a greater impact of the PVE. SR degradation due to target motion is much more difficult to codify than degradation due to physical, instrumental and computational effects. In fact, motion cannot be predicted in advance, since it depends on the specific patient and on the specific anatomical district [89, 90]. Organ motion is one of the main reasons why most of the PVE correction techniques were originally developed for neurological applications. In fact, in brain studies, the only movements that could arise were voluntary/involuntary movements of the head which can be modelled and compensated for with simple translational and/or rotational transformations [91]. The motion of districts other than the head (e.g. heart, pancreas etc.) cannot be considered rigid. In fact, the motion induced by respiration and heart beat consists not only of displacement of the organ under examination, but also in its deformation (periodic modifications in the size and shape of the organ).
Furthermore, PET image degradation is due not only to the blurring effect caused by the periodic spatial displacements of the organs, but also to the spatial mismatch between PET and anatomical data when anatomical data are used for PET data correction (e.g. attenuation correction, recovery of SR, PVE compensation). Since acquisition of anatomical data is not generally performed simultaneously with PET data acquisition and, furthermore, different acquisition protocols are used, spatial mismatches are likely to arise.
A typical solution to the problem of motion in cardiac and oncological applications is to perform a cardiac and/or respiratory gating of the data and to reconstruct individual PET-gated datasets with the corresponding matched anatomical images. Unfortunately, gating techniques lead to images with high noise as the count statistics in each gate decrease proportionally with the number of gates [92]. The problem becomes extremely severe when double gating techniques are used in attempts to compensate for both respiratory and cardiac motion [93, 94].
Therefore, more advanced approaches to PET imaging, moving beyond conventional gating and aiming to use all the collected events to generate a single high count statistics motion-corrected image set are under development. Two different approaches have been considered: (1) post-reconstruction registration by rigid or elastic transformation and summation of the independently reconstructed gated images (e.g. [95, 96]); (2) incorporation of the motion information directly within the reconstruction algorithm [97–99].
Motion compensation, combined with PVE correction, is the strategy needed to truly improve image quality and obtain accurate quantitative PET data (Fig. 8). Different groups have recently proposed methods for such double (motion and PVE) compensation [100–103]. For instance, a very interesting approach was recently proposed by Petibon et al. [103], which involved simultaneous acquisition of cardiac PET–MR data followed by reconstruction of the PET images accounting for non-rigid cardiac motion and the limited PSF of the scanner. In particular, an accurate non-rigid model of cardiac wall motion was measured by means of MR tagging sequences and then incorporated into the reconstruction of PET images. A motion ‘deblurring’ and a PSF compensation were therefore performed during the reconstruction process using all the available PET counts. The proposed methods have been evaluated in a beating non-rigid cardiac phantom. The results showed that tagged MR-based motion correction yielded an improvement in defect/myocardium contrast recovery as compared with motion-uncorrected studies. Likewise, lesion detectability improved with MR-based motion compensation as compared with gating and no motion correction and made it possible to distinguish non-transmural from transmural defects.
Clinical impact of PET quantification
Today, PET is widely used for the non-invasive in vivo assessment of patient-specific features of disease, with the aim of optimising the diagnostic and therapeutic management of the individual patient within a “personalised medicine” framework. The possibility of extracting, from PET images, specific indexes ascribable to disease status or to therapy response, and associating them with known prognostic factors or survival endpoints, opens up new perspectives on the role of PET in translating novel biomarkers from the bench to clinical use.
In cancer studies, a potential prognostic role for PET is acknowledged, based on the fact that glucose metabolism in primitive tumour lesions correlates with different biological cancer features (e.g. histological grade) with proven prognostic value. Several candidate PET biomarkers, based on PET data quantification, have been proposed, e.g. metabolic tumour volume (MTV), standardised uptake value (SUV), and total lesion glycolysis (TLG). Such biomarkers were found to be higher in patients with poor prognostic features, and thus to be helpful in determining the best candidates for baseline staging [e.g. 104]. However, the accuracy and reproducibility of PET biomarkers are major issues, and the impact of the PVE needs to be taken in consideration in this regard. Different approaches have been proposed to overcome the PVE problem. The maximum SUV value in the lesion (SUVmax) has been considered, as it has been found to be less affected by PVE than mean SUV. However, SUVmax is more sensitive to statistical fluctuations, and this sometimes leads to an overestimation of the true value. In some studies, only large tumours with small PVE were considered (e.g. anatomical dimension >2 cm) (e.g. [104]). More recently, standardised added metabolic activity (SAM) was proposed as a PVE-free biomarker of total lesion glycolysis [105].
The impact of PVE correction on the prognostic and predictive role of quantitative PET in cancer studies has been investigated by comparing PET biomarkers with or without PVE correction (Fig. 9). PVE correction has been shown to improve the statistical significance of correlations between PET biomarkers and biological prognostic indexes or survival endpoints [106–111]. Common findings suggest that PVE correction is required if PET biomarkers are used to stratify patients based on a cut-off value. When the SUV is used in diagnostic applications, i.e. to differentiate benign from malignant tumour based on an absolute cut-off value of SUV, definition of the latter should account for the PVE, otherwise it may prove inappropriate. Some studies have demonstrated that quantitative PET has a predictive role in therapy response. In oesophageal cancer, TLG appeared to be a predictive factor of concomitant radio-chemotherapy response with a significantly higher predictive value than SUV [107, 108]. In breast cancer, the reduction of TLG after neoadjuvant chemotherapy cycles was found to predict histopathological tumour response with higher accuracy than SUV, especially for ER-positive/HER2-negative breast cancer [109]. In non small cell lung cancer patients with tumours of considerable size who were submitted to curative resection, SUVmax was found to predict outcome even without PVE correction [110]. In head and neck cancer, when patients were submitted to a radical radiotherapy treatment, MTV and TLG (with and without PVE correction) were able to predict patient outcome, considering local and distant disease control endpoints [local recurrence-free survival (LRFS), distant metastasis-free survival, and disease-free survival (DFS)]. SUVmean was found to be predictive of LRFS and DFS and PVE correction was shown to increase the statistical significance of correlations [111].
As regards the role of PET in therapy monitoring during follow-up, Stefano et al. [112] showed that PVE strongly affected classification of therapy response of oncological lesions when the EORTC or PERCIST classification was used. Hoetjes et al. [11] measured SUV variation in patients with breast cancer after the first cycle of chemotherapy and found that SUVs showed an average reduction of 31 % in PVE-uncorrected images vs. 26 % in PVE-corrected ones. This suggests that if SUV was estimated after a chemotherapy session without taking the PVE into account, the resulting measurement of tumour metabolism might show a more pronounced decrease compared with its true status. Maisonobe et al. [113] showed that it was not necessary to apply PVE correction to SUV for predicting tumour response in patients with metastatic colorectal cancer treated with chemotherapy. In paediatric lymphoma patients, TLG appeared to be a useful quantitative parameter for the assessment of treatment response [114].
The PVE affects patterns of disease that may be of interest when investigating neurological biomarkers. An essential point when estimating neurological local indexes with PET is the ability to distinguish changes in radioactivity distribution due to the PVE from true changes in tissue function. For example, in patients with AD, cerebral cortex atrophy can lead to changes in PET local quantification which can be overestimated as a pure consequence of PVE. The importance of PVE correction in PET brain studies has been extensively described, e.g. by Thomas et al. [72], who showed a higher uptake in brain white matter (e.g. for 18F-flutemetamol) in patients with AD as a direct consequence of PVE. In other words, after PVE correction, no significant difference was found between AD patients and healthy controls. In the case of PET brain studies, as already described, a VB PVE correction algorithm was found to be particularly suitable and effective for quantifying metabolism in white matter regions at a voxel level and in correcting the spill-out from the white matter signal (e.g. in the ventricles), thus allowing accurate measurement of local biomarkers. More recent MRI-guided PVE compensation methods are emerging as very powerful tools for the accurate estimation of PET radioactivity concentration in the brain, also in consideration of the recent and future availability of hybrid PET/MRI systems.
However, the accuracy of anatomically guided PVE correction algorithms in PET brain images is largely affected by the performance of MRI segmentation algorithms partitioning the brain into its main classes [115, 116]. PVE becomes even more important when investigating biomarkers of neurodegenerative diseases with PET ligands. Thomas et al. [72], showed that special care should be taken when applying PVE correction to amyloid PET images. Assumptions made in existing PVE compensation strategies can in fact induce biases that could lead to erroneous inferences about the radiotracer uptake in certain brain structures and PVE compensation techniques must account for within-compartment variability to reduce errors of this kind. A significant bias in kinetic parameters was also evident in cerebral dynamic studies [117] as an effect of PVE correction. Performance of kinetic modelling from PET images processed with several PVE correction methods has been assessed on different brain PET radiotracer studies. The results indicate that the choice of PVE compensation method can significantly impact on the kinetic parameter estimation, and thus have consequences on the estimation of biomarkers deriving from the kinetic modelling.
As already discussed, the PVE and cardiac motion are two of the main image degradation effects in cardiac PET. Once motion is compensated for, the PVE introduces a systematic underestimation bias in measured myocardial radiotracer uptake, given the reduced thickness of the myocardial wall. Limited resolution is the cause of major signal cross-contamination between the cardiac chamber, regions with perfusion defects and normal myocardial tissue, which precludes accurate quantification of candidate biomarkers of disease. Newly available simultaneous PET/MRI scanners offer new possibilities in cardiac imaging as MRI can assess wall contractility while collecting PET perfusion data. Inflammation has been found to be a major risk factor for atherosclerotic plaque rupture and clinical events, especially when considering cardiac PET biomarkers. Previous studies have shown that PET 18F-FDG uptake in plaque correlates with macrophage content. However, the PVE can affect the measurement of plaque uptake. Izquierdo-Garcia et al. [118] demonstrated that vessel wall-to-blood ratio correlated better with the Patlak constant value than with the SUV, and showed a stronger correlation when PVE compensation was accounted for. Generally speaking, in cardiac disease, PVE correction was not found to be mandatory for qualitative clinical assessment of glucose metabolism in the vessel wall of abdominal aortic aneurysm patients but necessary to establish quantitative cut-off values to stratify patients for aneurysm repair [119].
Conclusions and future directions
The challenge for the next generation of in vivo molecular imaging systems is the detection and quantification of biological processes in living subjects at the cellular and sub-cellular levels. The ability of PET to obtain in vivo quantitative measurements of the distribution of a tracer in the human body places it at the forefront of molecular imaging techniques.
One particular strength of PET is its high sensitivity, given that current generation PET scanners are able to detect amounts of radiotracer even at picomolecular level. Conversely, spatial resolution unfortunately represents a weakness of PET, especially when compared with other imaging modalities (e.g. CT, MRI), resulting in poor image quality and in biases in image quantification.
To improve the performances of PET systems, future technological development should be directed towards overcoming limited spatial resolution, through improvements in the architecture of PET scanners, including the development of new PET detectors (e.g. new crystals, new photodetectors, novel designs of layered crystals providing depth-of-interaction information), signal processing methods and devices, and acquisition, reconstruction and post-processing methods. Improvements in spatial resolution could be achieved by optimising the design of the PET scanner to specific applications, e.g. brain-, breast-, prostate-dedicated tomographs. Another area of technological evolution is that of integrated scanners and, in particular, fully integrated PET/MRI systems, since MRI provides high-resolution anatomical information with excellent soft-tissue contrast without increasing the radiation dose to the patient. The simultaneous acquisition and fusion of PET and MRI yield spatially co-registered high-resolution and highly sensitive molecular images. Furthermore, motion correction can be directly accounted for within the PET reconstruction process, without loss of signal-to-noise ratio. The possibility of performing simultaneous motion correction and spatial resolution recovery may allow PET systems to reach their intrinsic limits for spatial resolution (2–3 mm) even in whole-body imaging.
To account for residual spatial resolution and its effect on image quantification, future developments should also be addressed at further improving and validating PVE compensation techniques. To provide the medical community with proper recommendations on their use, in-depth validation and cross-comparison studies should be performed. Advantages and limitations of the currently available methods should be considered with respect to the specific clinical applications. To facilitate the use and diffusion of PVE compensation techniques, implementation embedded into commercial software should be available.
PVE correction methods performed within the reconstruction process would be preferable for their easier clinical use. Techniques requiring anatomical information (CT, MRI) would be suitable when using integrated PET/CT or PET/MRI scanners, and appear particularly attractive for future molecular imaging scenarios.
Furthermore, the effect of compensation methods on image noise should be assessed and new strategies for noise reduction (e.g. regularisation) should be envisaged.
The availability of quantification methods, accurate and easy to implement in clinical settings, is crucial when PET is used for the extraction and quantification of biomarkers. In cardiological and oncological applications, characterised by local abnormalities in radiotracer uptake due to localised effects of the disease, region-based PVE correction methods may be appropriate for biomarker quantification. In brain studies, voxel-based PVE correction algorithms, accounting for spill-out effects from white matter, would be particularly effective in quantifying spatial patterns at a voxel level and/or allowing local biomarkers to be measured accurately.
The availability of practical, accurate and reproducible biomarker quantification methods could open new scenarios for the use of PET in the context of the new concept of personalised medicine. Novel PET predictive biomarkers, in helping to assign patients to optimal therapeutic approaches and increase the appropriateness of interventions, have the potential to help reduce health care costs. New PET prognostic biomarkers, on the other hand, will make it possible to predict the natural course of a specific disease and different outcomes. 18F-FDG PET, moving towards more personalised diagnostic and therapeutic patient management, is taking the first steps in this direction. Medical decisions will be helped by knowledge of the molecular characteristics of a given disease in the individual patient, measured by PET with 18F-FDG and ideally other disease-specific radiotracers, combined with the patient’s clinical data.
In the near future, quantitative PET and molecular imaging, being predictive, personalised, and contributing to medical decisions in individual patient management, really could change from a reactive to a proactive discipline.
References
Vallabhajosula S, Solnes L, Vallabhajosula B (2011) A broad overview of positron emission tomography radiopharmaceuticals and clinical applications: what is new? Semin Nucl Med 41:246–264
Lewellen TK (2010) The challenge of detector designs for PET. Am J Roentgenol 195:301–309
Levin Klausen T, Høgild Keller S, VinterOlesen O, Aznar M, Andersen FL (2012) Innovations in PET/CT. Q J Nucl Med Mol Imaging 56:268–279
Torigian DA, Zaidi H, Kwee TC, Saboury B, Udupa JK, Cho ZH, Alavi A (2013) PET/MR imaging: technical aspects and potential clinical applications. Radiology 267:26–44
Strobel K, Exner UE, Stumpe KDM, Hany TF, Bode B, Mende K, Veit-Haibach P, von Schulthess GK, Hodler J (2008) The additional value of CT images interpretation in the differential diagnosis of benign vs malignant primary bone lesions with 18F-FDG-PET/CT. Eur J Nucl Med Mol Imaging 35:2000–2008
Shankar LK, Hoffman JM, Bacharach S, Graham MM, Karp J, Lammertsma AA, Larson S, Mankoff DA, Siegel BA, Van den Abbeele A, Yap J, Sullivan D, National Cancer Institute (2006) Consensus recommendations for the use of 18F-FDG PET as an indicator of therapeutic response in patients in National Cancer Institute trials. J Nucl Med 47:901–903
Schmidt M, Bollschwieler E, Dietlein M, Mönig SP, Kobe C, Vallboehmer D, Eschner W, Hölscher A, Schicha H (2009) Mean and maximum standardized uptake values in [18F]FDG-PET for assessment of histopathological response in oesophageal squamous cell carcinoma or adenocarcinoma after radiochemotherapy. Eur J Nucl Med 36:735–744
Soret M, Bacharach SL, Buvat I (2007) Partial-volume effect in PET tumor imaging. J Nucl Med 48:932–945
Rousset OG, Zaidi H (2006) Correction for partial volume effects in emission tomography. In: Zaidi H (ed) Quantitative analysis in nuclear medicine imaging. Springer, New York, pp 236–271
Rousset O, Rahmim A, Alavi A, Zaidi H (2007) Partial volume correction strategies in PET. PET Clinics 2:235–249
Hoetjes NJ, van Velden FH, Hoekstra OS, Hoekstra CJ, Krak NC, Lammertsma AA, Boellaard R (2010) Partial volume correction strategies for quantitative FDG PET in oncology. Eur J Nucl Med Mol Imaging 37:1679–1687
Erlandsson K, Buvat I, Pretorius PH, Thomas BA, Hutton BF (2012) A review of partial volume correction techniques for emission tomography and their applications in neurology, cardiology and oncology. Phys Med Biol 57:R119–R159
Rahmim A, Qi J, Sossi V (2013) Resolution modeling in PET imaging: theory, practice, benefits and pitfalls. Med Phys 40:064301. doi:10.1118/1.4800806
Daube-Witherspoon ME, Karp JS, Casey ME, Di Filippo FP, Hines H, Muehllehner G, Simcic V, Stearns CW, Adam LE, Kohlmyer S, Sossi V (2002) PET performance measurements using the NEMA NU 2-2001 standard. J Nucl Med 43:1398–1409
Kotasidis FA, Matthews JC, Angelis GI, Noonan PJ, Jackson A, Price P, Lionheart WR, Reader AJ (2011) Single scan parameterization of space-variant point spread functions in image space via a printed array. The impact for two PET/CT scanners. Phys Med Biol 56:2917–2942
Panin VY, Kehren F, Michel C, Casey M (2006) Fully 3-D PET reconstruction with system matrix derived from point source measurements. IEEE Trans Med Imaging 25:907–921
Alessio AM, Stearns CW, Tong S, Ross SG, Kohlmyer S, Ganin A, Kinahan PE (2010) Application and evaluation of a measured spatially variant system model for PET image reconstruction. IEEE Trans Med Imaging 29:938–949
Kotasidis FA, Angelis GI, Anton-Rodriguez J, Matthews JC, Andrew J, Reader AJ, Zaidi H (2014) Isotope specific resolution recovery image reconstruction in high resolution PET imaging. Med Phys 41:052503. doi:10.1118/1.4870985
Hoffman EJ, Huang SC, Phelps ME (1979) Quantitation in positron emission computed tomography 1: effect of object size. J Comput Assist Tomogr 3:299–308
Hoffman EJ, Huang SC, Plummer D, Phelps ME (1982) Quantitation in positron emission computed tomography 6: effect of non uniform resolution. J Comput Assist Tomogr 6:987–999
Kessler RM, Ellis JR, Eden M (1984) Analysis of emission tomographic scan data: limitations imposed by resolution and background. J Comput Assist Tomogr 8:514–522
Weber WA, Ziegler SI, Thödtmann R, Hanauske AR, Schwaiger M (1999) Reproducibility of metabolic measurements in malignant tumors using FDG PET. J Nucl Med 40:1771–1777
Srinivas SM, Dhurairaj T, Basu S, Bural G, Surti S, Alavi A (2009) A recovery coefficient method for partial volume correction of PET images. Ann Nucl Med 23:341–348
Gallivanone F, Stefano A, Canevari C, Gianolli L, Messa C, Gilardi MC, Castiglioni I (2011) PVE correction in PET-CT whole-body oncological studies from PVE-affected images. IEEE Trans Nucl Sci 58:736–747
Gallivanone F, Canevari C, Gianolli L, Salvatore C, Della Rosa P, Gilardi MC, Castiglioni I (2013) A partial volume effect correction tailored for 18F-FDG-PET oncological studies. Biomed Res Int 780458. doi:10.1155/2013/780458
Huesman RH (1984) A new fast algorithm for the evaluation of regions of interest and statistical uncertainty in computed tomography. Phys Med Biol 29:543–552
Muzic RF Jr, Chen CH, Nelson AD (1998) A method to correct for scatter, spill over, and partial volume effects in region of interest analysis in PET. IEEE Trans Med Imaging 17:202–213
Carson RE (1986) A maximum likelihood method for region-of-interest evaluation in emission tomography. J Comput Assist Tomogr 10:654–663
Formiconi AR (1993) Least squares algorithm for region-of-interest evaluation in emission tomography. IEEE Trans Med Imaging 12:90–100
Vanzi E, De Cristofaro MT, Ramat S, Sotgia B, Mascalchi M, Formiconi AR (2007) A direct ROI quantification method for inherent PVE correction: accuracy assessment in striatal SPECT measurements. Eur J Nucl Med Mol Imaging 34:1480–1489
Moore SC, Southekal S, Park MA, McQuaid SJ, Kijewski MF, Müller SP (2012) Improved regional activity quantitation in nuclear medicine using a new approach to correct for tissue partial volume and spill over effects. IEEE Trans Med Imaging 31:405–416
Rousset OG, Ma Y, Evans AC (1998) Correction for partial volume effects in PET: principle and validation. J Nucl Med 39:904–911
Du Y, Tsui BM, Frey EC (2005) Partial volume effect compensation for quantitative brain SPECT imaging. IEEE Trans Med Imaging 24:969–976
Sattarivand M, Kusano M, Poon I, Caldwell C (2012) Symmetric geometric transfer matrix partial volume correction for PET imaging: principle, validation, and robustness. Phys Med Biol 57:7101–7116
Reader AJ, Zaidi H (2007) Advances in PET image reconstruction. PET Clin 2:173–190
Leahy RM, Qi J (2000) Statistical approaches in quantitative positron emission tomography. Stat Comput 10:147–165
Bai B, Li Q, Leahy RM (2013) Magnetic resonance-guided positron emission tomography image reconstruction. Semin Nucl Med 43:30–44
Reader AJ, Julyan PJ, Williams H, Hastings DL, Zweit J (2003) EM algorithm system modeling by image-space techniques for PET reconstruction. IEEE Trans Nucl Sci 50:1392–1397
Tong S, Alessio A, Thielemans K, Stearns C, Ross S, Kinahan P (2011) Edge artifacts in PSF-based PET image reconstruction: characterization and mitigation. J Nucl Med 52(Supplement 1):106
Sureau FC, Reader AJ, Comtat C, Leroy C, Ribeiro MJ, Buvat I, Trebossen R (2008) Impact of image-space resolution modeling for studies with the high-resolution research tomography. J Nucl Med 49:1000–1008
Bowen SL, Byars LG, Michel CJ, Chonde DB, Catana C (2013) Influence of the partial volume correction method on (18)F-fluorodeoxyglucose brain kinetic modelling from dynamic PET images reconstructed with resolution model based OSEM. Phys Med Biol 58:7081–7106
Lasnon C, Hicks RJ, Beauregard JM, Milner A, Paciencia M, Guizard AV, Bardet S, Gervais R, Lemoel G, Zalcman G, Aide N (2012) Impact of point spread function reconstruction on thoracic lymph node staging with 18F-FDG PET/CT in non-small cell lung cancer. Clin Nucl Med 37:971–976
Orlandini LC, Betti M, Fulcheri C, Dona M, Fisicaro D, Castagnoli A (2013) Improvement in clinical evaluation of PET/CT images with high resolution algorithms. Q J Nucl Med Mol Imaging 57:201–206
Andersen FL, Klausen TL, Loft A, Beyer T, Holm S (2013) Clinical evaluation of PET image reconstruction using a spatial resolution model. Eur J Radiol 82:862–869
Conti M (2009) State of the art and challenges of time-of-flight PET. Phys Med 25:1–11
El Fakhri G, Surti S, Trott CM, Scheuermann J, Karp JS (2011) Improvement in lesion detection with whole-body oncologic time-of-flight PET. J Nucl Med 52:347–353
Kadrmas DJ, Casey ME, Conti M, Jakoby BW, Lois C, Townsend DW (2009) Impact of time-of-flight on PET tumor detection. J Nucl Med 50:1315–1323
Schaefferkoetter J, Casey M, Townsend DW, El Fakhri G (2013) Clinical impact of time-of-flight and point response modeling in PET reconstructions: a lesion detection study. Phys Med Biol 58:1465–1478
Prieto E, Domínguez-Prado I, García-Velloso MJ, Peñuelas I, Richter JÁ, Martí-Climent JM (2013) Impact of time-of-flight and point-spread-function in SUV quantification for oncological PET. Clin Nucl Med 38:103–109
Sibarita JB (2005) Deconvolution microscopy. Adv Biochem Eng Biotechnol 95:201–243
Teo BK, Seo Y, Bacharach SL, Carrasquillo JA, Libutti SK, Shukla H, Hasegawa BH, Hawkins RA, Franc BL (2007) Partial-volume correction in PET: validation of an iterative post-reconstruction method with phantom and patient data. J Nucl Med 48:802–810
Tohka J, Reilhac A (2008) Deconvolution-based partial volume correction in Raclopride-PET and Monte Carlo comparison to MR-based method. Neuroimage 39:1570–1584
Kirov AS, Piao JZ, Schmidtlein CR (2008) Partial volume effect correction in PET using regularized iterative deconvolution with variance control based on local topology. Phys Med Biol 53:2577–2591
Boussion N, Cheze Le Rest C, Hatt M, Visvikis D (2009) Incorporation of wavelet-based denoising in iterative deconvolution for partial volume correction in whole-body PET imaging. Eur J Nucl Med Mol Imaging 36:1064–1075
Rizzo G, Castiglioni I, Russo G, Tana MG, Dell’Acqua F, Gilardi MC, Fazio F, Cerutti S (2007) Using deconvolution to improve PET spatial resolution in OSEM iterative reconstruction. Methods Inf Med 46:231–235
Boussion N, Hatt M, Lamare F, Bizais Y, Turzo A, Cheze-Le Rest C, Visvikis D (2006) A multiresolution image based approach for correction of partial volume effects in emission tomography. Phys Med Biol 51:1857–1876
Le Pogam A, Hatt M, Descourt P, Boussion N, Tsoumpas C, Turkheimer FE, Prunier-Aesch C, Baulieu JL, Guilloteau D, Visvikis D (2011) Evaluation of a 3D local multiresolution algorithm for the correction of partial volume effects in positron emission tomography. Med Phys 38:4920–4923
Shidahara M, Tsoumpas C, Hammers A, Boussion N, Visvikis D, Suhara T, Kanno I, Turkheimer FE (2009) Functional and structural synergy for resolution recovery and partial volume correction in brain PET. Neuroimage 44:340–348
Videen TO, Perlmutter JS, Mintun MA, Raichle ME (1988) Regional correction of positron emission tomography data for the effects of cerebral atrophy. J Cereb Blood Flow Metab 8:662–670
Meltzer CC, Leal JP, Mayberg HS, Wagner HN Jr, Frost JJ (1990) Correction of PET data for partial volume effects in human cerebral cortex by MR imaging. J Comput Assist Tomogr 14:561–570
Müller-Gärtner HW, Links JM, Prince JL, Bryan RN, McVeigh E, Leal JP, Davatzikos C, Frost JJ (1992) Measurement of radiotracer concentration in brain gray matter using positron emission tomography: MRI-based correction for partial volume effects. J Cereb Blood Flow Metab 12:571–583
Meltzer CC, Cantwell MN, Greer PJ, Ben-Eliezer D, Smith G, Frank G, Kaye WH, Houck PR, Price JC (2000) Does cerebral blood flow decline in healthy aging? A PET study with partial-volume correction. J Nucl Med 41:1842–1848
Yanase D, Matsunari I, Yajima K, Chen W, Fujikawa A, Nishimura S, Matsuda H, Yamada M (2005) Brain FDG PET study of normal aging in Japanese: effect of atrophy correction. Eur J Nucl Med Mol Imaging 32:794–805
Ibáñez V, Pietrini P, Furey ML, Alexander GE, Millet P, Bokde AL, Teichberg D, Schapiro MB, Horwitz B, Rapoport SI (2004) Resting state brain glucose metabolism is not reduced in normotensive healthy men during aging, after correction for brain atrophy. Brain Res Bull 63:147–154
Ibáñez V, Pietrini P, Alexander GE, Furey ML, Teichberg D, Rajapakse JC, Rapoport SI, Schapiro MB, Horwitz B (1998) Regional glucose metabolic abnormalities are not the result of atrophy in Alzheimer’s disease. Neurology 50:1585–1593
Meltzer CC, Zubieta JK, Links JM, Brakeman P, Stumpf MJ, Frost JJ (1996) MR-based correction of brain PET measurements for heterogeneous gray matter radioactivity distribution. J Cereb Blood Flow Metab 16:650–658
Du Y, Madar I, Stumpf MJ, Rong X, Fung GS, Frey EC (2013) Compensation for spill-in and spill-out partial volume effects in cardiac PET imaging. J Nucl Cardiol 20:84–98
Rousset OG, Ma Y, Wong DF, Evans AC (1998) Pixel-versus region-based partial volume correction in PET. In: Carson RE, Herscovitch P, Daube-Witherspoon M (eds) Quantitative functional brain imaging with positron emission tomography. Academic Press, San Diego, pp 67–75
Quarantelli M, Berkouk K, Prinster A, Landeau B, Svarer C, Balkay L, Alfano B, Brunetti A, Baron JC, Salvatore M (2004) Integrated software for the analysis of brain PET/SPECT studies with partial-volume-effect correction. J Nucl Med 45:192–201
Alfano B, Quarantelli M, Comerci M, Brunetti A, Salvatore M (2004) A new method for voxel-based partial volume effect correction. IBB HBM Poster # TH360
Yang J, Huang SC, Mega M, Lin KP, Toga AW, Small GW, Phelps ME (1996) Investigation of partial volume correction methods for brain FDG PET studies. IEEE Trans Nucl Sci 43:3322–3327
Thomas BA, Erlandsson K, Modat M, Thurfjell L, Vandenberghe R, Ourselin S, Hutton BF (2011) The importance of appropriate partial volume correction for PET quantification in Alzheimer’s disease. Eur J Nucl Med Mol Imaging 38:1104–11019
Erlandsson K, Hutton BF (2010) Partial volume correction in SPECT using anatomical information and iterative FBP. Tsinghua Sci Technol 15:50–55
Erlandsson K, Thomas B, Dickson J, Hutton B F (2010) Evaluation of an OSEM-based PVC method for SPECT with clinical data. In: Nuclear Science Symposium and Medical Imaging Conference Record pp 2686–2689
Harri M, Mika T, Jussi H, Nevalainen OS, Jarmo H (2007) Evaluation of partial volume effect correction methods for brain positron emission tomography: quantification and reproducibility. J Med Phys 32:108–117
Aston JA, Cunningham VJ, Asselin MC, Hammers A, Evans AC, Gunn RN (2002) Positron emission tomography partial volume correction: estimation and algorithms. J Cereb Blood Flow Metab 22:1019–1034
Carson RE (2003) Tracer kinetic modeling in PET. Chapter 6 from Valk PE, Bailey DL, Townsend DW, Maisey MN. Positron emission tomography: basic science and clinical practice. Springer, London Ltd 2003, pp 147–179
Bentourkia M, Zaidi H (2007) Positron emission tomography. Tracer kinetic modeling in PET. PET Clin 2:267–277
Karakatsanis NA, Lodge MA, Zhou Y, Wahl RL, Rahmim A (2013) Dynamic whole-body PET parametric imaging: II. Task-oriented statistical estimation. Phys Med Biol 58:7419–7445
Litton JE (1997) Input function in PET brain studies using MR-defined arteries. J Comput Assist Tomogr 21:907–909
Chen K, Bandy D, Reiman E, Huang SC, Lawson M, Feng D, Yun LS, Palant A (1998) Non invasive quantification of the cerebral metabolic rate for glucose using positron emission tomography, 18F-fluoro-2-deoxyglucose, the Patlak method, and an image-derived input function. J Cereb Blood Flow Metab 18:716–723
Mourik JE, Lubberink M, Klumpers UM, Comans EF, Lammertsma AA, Boellaard R (2008) Partial volume corrected image derived input functions for dynamic PET brain studies: methodology and validation for [11C]flumazenil. Neuroimage 39:1041–1050
Croteau E, Lavallée E, Labbe SM, Hubert L, Pifferi F, Rousseau JA, Cunnane SC, Carpentier AC, Lecomte R, Bénard F (2010) Image-derived input function in dynamic human PET/CT: methodology and validation with 11C-acetate and 18F-fluorothioheptadecanoic acid in muscle and 18F-fluorodeoxyglucose in brain. Eur J Nucl Med Mol Imaging 37:1539–1550
Hackett SL, Liu D, Chalkidou A, Marsden P, Landau D, Fenwick JD (2013) Estimation of input functions from dynamic [18F]FLT PET studies of the head and neck with correction for partial volume effects. EJNMMI Res 3:84. doi:10.1186/2191-219X-3-84
Zanotti-Fregonara P, Chen K, Liow JS, Fujita M, Innis RB (2011) Image-derived input function for brain PET studies: many challenges and few opportunities. J Cereb Blood Flow Metab 31:1986–1998
Hutchins GD, Caraher JM, Raylman RR (1992) A region of interest strategy for minimizing resolution distortions in quantitative myocardial studies. J Nucl Med 33:1243–1250
Iida H, Rhodes CG, de Silva R, Yamamoto Y, Araujo LI, Maseri A, Jones T (1991) Myocardial tissue fraction-correction for partial volume effects and measure of tissue viability. J Nucl Med 32:2169–2175
Rahmim A, Tang J, Zaidi H (2009) Four-dimensional (4D) image reconstruction strategies in dynamic PET: beyond conventional independent frame reconstruction. Med Phys 36:3654–3670
Bettinardi V, Picchio M, Di Muzio N, Gilardi MC (2012) Motion management in positron emission tomography/computed tomography for radiation treatment planning. Semin Nucl Med 42:289–307
Pépin A, Daouk J, Bailly P, Hapdey S, Meyer ME (2012) Management of respiratory motion in PET/computed tomography: the state of the art. Nucl Med Commun 35:113–122
Rahmim A (2005) Advanced motion correction methods in PET. Iran J Nucl Med 13:2–17
Ter-Pogossian MM, Bergman SR, Sobel BE (1982) Influence of cardiac and respiratory motion on tomographic reconstructions of the heart: implications for quantitative nuclear cardiology. J Comput Assist Tomogr 6:1148–1155
Martinez-Möller A, Zikic D, Botnar RM, Bundschuh RA, Howe W, Ziegler SI, Navab N, Schwaiger M, Nekolla SG (2007) Dual cardiac-respiratory gated PET: implementation and results from a feasibility study. Eur J Nucl Med Mol Imaging 34:1447–1454
Teräs M, Kokki T, Durand-Schaefer N, Noponen T, Pietilä M, Kiss J, Hoppela E, Sipilä HT, Knuuti J (2010) Dual-gated cardiac PET-clinical feasibility study. Eur J Nucl Med Mol Imaging 37:505–516
Slomka PJ, Hidetaka Nishina H, Berman DS, Kang X, Akincioglu C, Friedman JD, Hayes SW, Aladl UE, Germano G (2004) “Motion-frozen” display and quantification of myocardial perfusion. J Nucl Med 45:1128–1134
Lamare F, Cresson T, Savean J, Cheze Le Rest C, Reader AJ, Visvikis D (2007) Respiratory motion correction for PET oncology applications using affine transformation of list mode data. Phys Med Biol 52:121–140
Qiao F, Pan T, Clark JW Jr, Mawlawi OR (2006) A motion-incorporated reconstruction method for gated PET studies. Phys Med Biol 51:3769–3783
Chun SY, Reese TG, Ouyang J, Guerin B, Catana C, Zhu X, Alpert NM, El Fakhri G (2012) MRI-based non rigid motion correction in simultaneous PET/MRI. J Nucl Med 53:1284–1291
Guérin B, Cho S, Chun SY, Zhu X, Alpert NM, El Fakhri G, Reese T, Catana C (2011) Non rigid PET motion compensation in the lower abdomen using simultaneous tagged-MRI and PET imaging. Med Phys 38:3025–3038
Apostolova I, Wiemker R, Paulus T, Kabus S, Dreilich T, van den Hoff J, Plotkin M, Mester J, Brenner W, Buchert R, Klutmann S (2010) Combined correction of recovery effect and motion blur for SUV quantification of solitary pulmonary nodules in FDG PET/CT. Eur Radiol 20:1868–1877
Chang G, Chang T, Pan T, Clark JW Jr, Mawlawi OR (2010) Joint correction of respiratory motion artefact and partial volume effect in lung/thoracic PET/CT imaging. Med Phys 37:6221–6232
Le Meunier L, Slomka PJ, Dey D, Ramesh A, Thomson LE, Hayes SW, Friedman JD, Cheng V, Germano G, Berman DS (2011) Motion frozen (18)F-FDG cardiac PET. J Nucl Cardiol 18:259–266
Petibon Y, Ouyang J, Zhu X, Huang C, Reese TG, Chun SY, Li Q, El Fakhri G (2013) Cardiac motion compensation and resolution modeling in simultaneous PET-MR: a cardiac lesion detection study. Phys Med Biol 58:2085–2102
Groheux D, Giacchetti S, Moretti JL, Porcher R, Espié M, Lehmann-Che J, de Roquancourt A, Hamy AS, Cuvier C, Vercellino L, Hindié E (2011) Correlation of high 18F-FDG uptake to clinical, pathological and biological prognostic factors in breast cancer. Eur J Nucl Med Mol Imaging 38:426–435
Mertens J, Dobbeleir A, Ham H, Yves D’Asseler Y, Goethals I, Van de Wiele C (2012) Standardized added metabolic activity (SAM): a partial volume independent marker of total lesion glycolysis in liver metastases. Eur J Nucl Med Mol Imaging 39:1441–1448
Gallivanone F, Canevari C, Sassi I, Zuber V, Marassi A, Gianolli L, Picchio M, Messa C, Gilardi MC, Castiglioni I (2014) Partial volume corrected 18F-FDG PET mean standardized uptake value correlates with prognostic factors in breast cancer. Q J Nucl Med Mol Imaging [Epub ahead of print]
Hatt M, Visvikis D, Pradier O, Cheze Le Rest C (2011) Baseline 18F-FDG PET image-derived parameters for therapy response prediction in oesophageal cancer. Eur J Nucl Med Mol Imaging 38:1595–1606
Hatt M, Le Pogam A, Visvikis D, Pradier O, Cheze Le Rest C (2012) Impact of partial-volume effect correction on the predictive and prognostic value of baseline 18F-FDG PET images in esophageal cancer. J Nucl Med 53:12–20
Hatt M, Groheux D, Martineau A, Espié M, Hindié E, Giacchetti S, de Roquancourt A, Visvikis D, Cheze-Le Rest C (2013) Comparison between 18F FDG PET image-derived indices for early prediction of response to neoadjuvant chemotherapy in breast cancer. J Nucl Med 54:341–349
Ohtaka K, Hida Y, Kaga K, Okamoto S, Shiga T, Tamaki N, Muto J, Nakada-Kubota R, Hirano S, Matsui Y (2013) Outcome analysis of 18F fluorodeoxyglucose positron–emission tomography in patients with lung cancer after partial volume correction. Anticancer Res 33:5193–5198
Picchio M, Kirienko M, Mapelil P, Dell’Oca I, Villa E, Gallivanone F, Gianolli L, Messa C, Castiglioni I (2014) Predictive value of pre-therapy 18F-FDG PET/CT for the outcome of 18F-FDG PET-guided radiotherapy in patients with head and neck cancer. Eur J Nucl Med Mol Imaging 41:21–31
Stefano A, Gallivanone F, Messa C, Gilardi MC, Castiglioni I (2014) Metabolic impact of partial volume correction of [18F]FDG PET-TC oncological studies on the assessment of tumor response to treatment. Q J Nucl Med Mol Imaging [Epub ahead of print]
Maisonobe JA, Garcia CA, Necib H, Vanderlinden B, Hendlisz A, Flamen P, Buvat I (2013) Comparison of PET metabolic indices for the early assessment of tumour response in metastatic colorectal cancer patients treated by polychemotherapy. Eur J Nucl Med Mol Imaging 40:166–1674
Sharma P, Gupta A, Patel C, Bakhshi S, Malhotra A, Kumar R (2012) Pediatric lymphoma: metabolic tumor burden as a quantitative index for treatment response evaluation. Ann Nucl Med 12:58–66
Zaidi H, Ruest T, Schoenahl F, Montandon ML (2006) Comparative assessment of statistical brain MR image segmentation algorithms and their impact on partial volume correction in PET. Neuroimage 32:1591–1607
Gutierrez D, Montandon ML, Assal F, Allaoua M, Ratib O, Lovblad KO, Zaidi H (2013) Anatomically guided voxel-based partial volume effect correction in brain PET: impact of MRI segmentation. Comp Med Imaging Graph 36:610–619
Rousset OG, Deep P, Kuwabara H, Evans AC, Gjedde AH, Cumming P (2000) Effect of partial volume correction on estimates of the influx and cerebral metabolism of 6-[(18)F]fluoro-l-dopa studied with PET in normal control and Parkinson’s disease subjects. Synapse 37:81–89
Izquierdo-Garcia D, Davies JR, Graves MJ, Rudd JH, Gillard JH, Weissberg PL, Fryer TD, Warburton EA (2009) Comparison of methods for magnetic resonance-guided [18-F]fluorodeoxyglucose positron emission tomography in human carotid arteries: reproducibility, partial volume correction, and correlation between methods. Stroke 40:86–93
Reeps C, Bundschuh RA, Pellisek J, Herz M, van Marwick S, Schwaiger M, Eckstein HH, Nekolla SG, Essler M (2013) Quantitative assessment of glucose metabolism in the vessel wall of abdominal aortic aneurysms: correlation with histology and role of partial volume correction. Int J Cardiovasc Imaging 29:505–512
Acknowledgments
This work was partly supported by the CNR Project PP.P01/Personalized Medicine 2013 and by the Project “MbMM–Basic methodologies for innovation in the diagnosis and therapy of multifactorial diseases” (signed 25/7/2013), Framework Agreement Lombardy Region-National Research Council of Italy (16/7/2012).
Conflict of interest
The authors, Valentino Bettinardi, Isabella Castiglioni, Elisabetta De Bernardi and Maria Carla Gilardi declare no conflict of interest.
Compliance with Ethics Guidelines
This article does not contain any studies with human or animal subjects performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Additional information
Color figures online at http://link.springer.com/article/10.1007/s40336-014-0066-y.
Rights and permissions
About this article
Cite this article
Bettinardi, V., Castiglioni, I., De Bernardi, E. et al. PET quantification: strategies for partial volume correction. Clin Transl Imaging 2, 199–218 (2014). https://doi.org/10.1007/s40336-014-0066-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40336-014-0066-y