-
PDF
- Split View
-
Views
-
Cite
Cite
Yukio Takeshita, Ryutaro Fujinaga, Changjiu Zhao, Akie Yanai, Koh Shinoda, Huntingtin-associated protein 1 (HAP1) interacts with androgen receptor (AR) and suppresses SBMA-mutant-AR-induced apoptosis, Human Molecular Genetics, Volume 15, Issue 15, 1 August 2006, Pages 2298–2312, https://doi.org/10.1093/hmg/ddl156
Close - Share Icon Share
Abstract
Huntingtin-associated protein 1 (HAP1), an interactor of huntingtin, has been known as an essential component of the stigmoid body (STB) and recently reported to play a protective role against neurodegeneration in Huntington's disease (HD). In the present study, subcellular association between HAP1 and androgen receptor (AR) with a long polyglutamine tract (polyQ) derived from spinal-and-bulbar-muscular-atrophy (SBMA) was examined using HEp-2 cells cotransfected with HAP1 and/or normal ARQ25, SBMA-mutant ARQ65 or deletion-mutant AR cDNAs. The results provided the first clear evidence that HAP1 interacts with AR through its ligand-binding domain in a polyQ-length-dependent manner and forms prominent inclusions sequestering polyQ-AR, and that addition of dihydrotestosterone reduces the association strength of HAP1 with ARQ25 more dramatically than that with ARQ65. Furthermore, SBMA-mutant-ARQ65-induced apoptosis was suppressed by cotransfection with HAP1. Our findings strongly suggest that HAP1/STB is relevant to polyQ-length-dependent modification on subcellular AR functions and critically involved in pathogenesis of not only HD but also SBMA as an important intrinsic neuroprotectant determining the threshold for cellular vulnerability to apoptosis. Taking together with previous reports that HAP1/STB is selectively expressed in the brain regions spared from degenerative targets in HD and SBMA, the current study might explain the region-specific occurrence of neurodegeneration in both diseases, shedding light on common aspects of their molecular pathological mechanism and yet-to-be-uncovered diagnostic or therapeutic applications for HD and SBMA patients.
INTRODUCTION
Over the past decade, a striking number of distinct, spherical-to-ovoidal cytoplasmic inclusions with unknown function, the stigmoid bodies (STBs), have been found prevalently in the limbic and hypothalamic regions of the rodent and primate brains ( 1–6 ). The STBs were originally detected by immunohistochemistry for human placental antigen X-P2 (hPAX-P2) and identified as non-membrane-bound inclusions (0.5–3 µm in diameter) with a granulo-fuzzy texture and moderate-to-low electron density ( 1 , 2 ). In 1998, transfection of huntingtin-associated protein 1 (HAP1)-cDNA into HEK293 was demonstrated to induce STBs ( 7 ). Subsequently, HAP1 was detected in the STB of the rat brain in light and electron microscopy ( 4 ). The distribution of HAP1mRNA expression in the rat ( 8 ) and mouse ( 9 ) brains corresponds to STB's distribution. Recent studies have shown that STB formation is inhibited in the brain of Hap1 (+/−) mice ( 5 , 6 ). All these data strongly suggest that HAP1 is an essential component of the STB in the neuronal cytoplasm in vivo and in vitro .
HAP1 was first identified by yeast two-hybrid screening ( 10 ) as an intraneuronal binding partner of huntingtin (Htt), a causative gene product in Huntington's disease (HD), which is an adult-onset autosomal-dominant polyglutamine (polyQ)-neurodegenerative disorder ( 11 ). Clinical symptoms of HD such as involuntary movements, dementia and other psychiatric disorders ( 12 , 13 ) involve selective atrophy of the striatum, thalamus and cerebral neocortex ( 14–16 ). Htt is, however, not specifically localized in the atrophic regions, but is widely expressed through the entire brain in rodents and humans ( 17 , 18 ). If HAP1 with a polyQ-length-dependent affinity to Htt ( 10 ) was distributed in regions affected in HD, it would help explain its region-specific pathogenesis, which seems inconsistent with the ubiquitous expression of Htt ( 10 , 19 ). But in situ hybridization of HAP1mRNA ( 9 ) indicated that HAP1 is preferentially expressed in the limbic and hypothalamic regions where neurodegeneration rarely occurs, and not in the striatum, thalamus and neocortex. Thus, we suggested that HAP1 plays a protective role against neurodegeneration rather than inducing neuronal death ( 9 ). Recently, it has been reported that in vitro transfection of HAP1 partially inhibits the activation of caspase-3 and suppresses apoptosis in polyQ-Htt-transfected HEK293 cells ( 6 ). This supports the ‘HAP1 protection hypothesis’ ( 9 ).
A physiological function of HAP1/STB in normal brains has, yet, to be determined. Distribution of HAP1, however, resembles that of androgen or estrogen receptors (AR, ER) ( 1 ); the STB coexists with ERα at a high frequency, particularly in the medial preoptic area, bed nucleus of the stria terminalis and medial amygdaloid nucleus ( 20 ). Intriguingly, the human AR gene has been reported to have a CAG-repeat motif near its 5′-terminus, as seen in Htt, being translated to AR protein with a polyQ sequence near the N-terminus ( 21 ). Furthermore, expansion of the triplet-CAG repeat of the AR gene also elicits another distinct polyQ-neurodegenerative disease, ‘spinal and bulbar muscular atrophy (SBMA)’ [Kennedy's disease or Kennedy-Alter-Sung syndrome (KAS)] ( 22–25 ). SBMA is an adult-onset X-linked disorder characterized by progressive proximal (bulbar and limb) muscle atrophy, weakness and fasciculations ( 22 , 23 , 26 , 27 ). The disease distribution does not rigidly follow such AR distribution; most of the forebrain regions rich in AR are spared. We have postulated that HAP1 also serves as a neuroprotectant against apoptosis brought about by the polyQ expansion of AR in SBMA pathogenesis.
In the present study, in order to obtain evidence for an interaction between HAP1 and AR in vitro , western blot, immunoprecipitations, and transfection of normal- or abnormally polyQ-expanded-AR, AR-deletion mutants and/or GFP-fused-HAP1 cDNAs into cultured HEp-2 cells have been used to examine: (1) whether or not HAP1 interacts with AR; (2) if so, whether the interaction is polyQ-length-dependent; (3) what domain of AR is essential for the association with HAP1; (4) whether or not HAP1 could inhibit apoptosis induced by extraordinarily expanded polyQ-AR in the presence of dihydrotestosterone (DHT).
RESULTS
Interaction of HAP1 with AR
Cotransfection of HAP1-cDNA and AR-cDNA
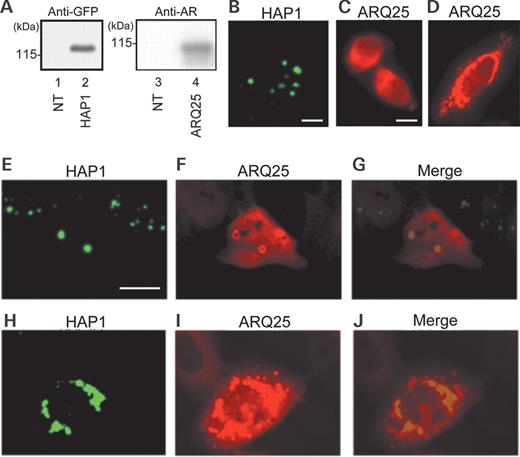
In order to evaluate the morphological interaction between HAP1 and AR, cDNAs of GFP-fused-HAP1 (GFP-HAP1) and FLAG-tagged AR with a normal length of poly-Q tract (ARQ25) were constructed and separately transfected into HEp-2 cells. Western blot analysis of the transfected cell lysates using anti-GFP- or anti-AR-antibody revealed a distinct band corresponding to the expected molecular weight of GFP-HAP1 or ARQ25, confirming successful expression of each molecule in the transfected HEp-2 cells (Fig. 1 A). By a fluorescence microscopy, most of the GFP-HAP1-transfected cultured cells were observed to form GFP-positive (0.5–5 µm in diameter) solitary inclusions in cytoplasm similar to the STBs as reported before ( 7 ). Indirect immunofluorescence cytochemistry with Alexa594-labeled anti-AR-antibody showed many ARQ25-transfected cells to express AR diffusely in cytoplasm and to a slight degree in the nucleus (Fig. 1 C). In addition, AR-immunopositive (AR-I) cytoplasmic inclusions or aggregations were expressed in 40% of the ARQ25-transfected cultured cells. The number of AR-I inclusions per cell varied; in some cells, a few solitary AR-I inclusions were observed in the cytoplasm, whereas, in others, large numbers assembled into a huge mass (Fig. 1 D).
Expression of HAP1 and/or AR after single- or cotransfection of cDNAs of GFP-HAP1 and FLAG-ARQ25 into HEp-2 cells in the control (DHT undetectable) medium. ( A ) Western blots with an anti-GFP (left) antibody or anti-AR (N20) (right) antibody for extracts (15 µg) from non-transfected (NT: lanes 1 and 3), GFP-HAP1-transfected (Lane 2) and ARQ25-transfected (Lane 4) cells. Molecular weight (kDa) is indicated in the left of each panel. ( B ) A fluorescent photomicrograph showing subcellular HAP1 expression (GFP: green) in HAP1-transfected cells. Note that HAP1 forms dot-like structures in the cytoplasm (bar=10 µm). ( C and D ) Fluorescent photomicrographs showing subcellular ARQ25 expression (Alexa594: red) in ARQ25-transfected cells. ARQ25 was detected with Alexa594-conjugated secondary antibodies following incubation with an anti-AR antibody (N20). Note that ARQ25 is diffusely present in the cytoplasm (and some are present in the nucleus) (C) or assembled into a huge mass of aggregations in perikarya (D) (bar=10 µm). ( E – J ) Fluorescent photomicrographs showing the subcellular expression of HAP1 (GFP: green) and ARQ25 (Alexa594: red) after cotransfection of their cDNAs. Note that merged images show the colocalization of HAP1 and ARQ25 as ring-shaped solitary cytoplasmic inclusions (E, F and G) or a huge mass of aggregations in perikarya (H, I and J) (bar=10 µm).
To evaluate the relationship between HAP1 and ARQ25, cDNAs of GFP-HAP1 and ARQ25 were cotransfected into HEp-2 cells. Fluorescence of GFP (indicating HAP1) and Alexa594 (marking ARQ25) were found to colocalize to solitary inclusions or huge aggregation clusters composed of smaller HAP1/AR inclusions (Figs 1 E–J). The solitary HAP1/AR double-positive structures resembled inclusions (or STB) seen after GFP-HAP1-single-transfection, while the large aggregates resembled the mass as detected in the perikarya after ARQ25-single-transfection. AR was prone to assembling in the periphery of both solitary inclusions and huge aggregation clusters (Fig. 1 G and J); in particular, it tended to show a ring-shaped appearance in the solitary inclusions.
Construction of SBMA-mutant AR with a 65-polyQ tract
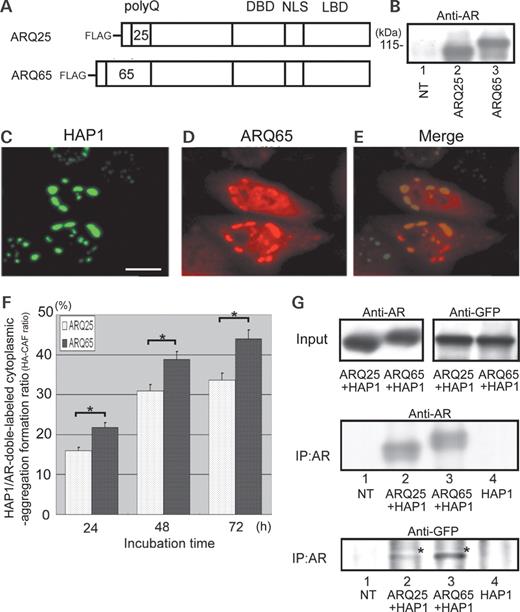
A FLAG-tagged-AR-cDNA with extraordinarily long CAG repeats derived from an SBMA patient ( 28 ), was made and transfected into HEp-2 cells. In western blot using anti-AR-antibody, the transfected cell lysates revealed a distinct band corresponding to the expected molecular weight of FLAG-tagged SBMA-mutant AR with 65-polyQ repeats (ARQ65), indicating ARQ65 expression in the transfected cells (Fig. 2 A and B). Immunofluorescence cytochemistry showed that the HAP1/AR-double-labeled cytoplasmic-aggregation formation (HA-CAF) occurs after cotransfection of cDNAs of GFP-HAP1 and ARQ65 into HEp-2 cells (Fig. 2 C, D and E).
PolyQ-length-dependent interaction between HAP1 and AR. ( A ) Diagrams of the primary structures of FLAG-tagged normal AR (ARQ25) and SBMA-derived AR (ARQ65). ( B ) Western blot with an anti-AR antibody (N20) for extracts (15 µg) from non-transfected (Lane 1), ARQ25-transfected (Lane 2) and ARQ65-transfected (Lane 3) cells. Molecular weight (kDa) is indicated in the left. ( C – E ) Fluorescent photomicrographs showing subcellular expression of HAP1 (GFP: green) and ARQ65 (Alexa594: red) after cotransfection of their cDNAs into HEp2-cells. Note that merged images show colocalization of HAP1 and ARQ65 (E) (bar=10 µm). ( F ) Bar graph comparing HA-CAF ratios between ARQ25 and ARQ65 in time course (24, 48 and 72 h) after the cotransfection into HEp-2 cells. Asterisks indicate the statistical significance of the difference in the HA-CAF ratio between ARQ65 and ARQ25 ( P <0.05). ( G ) Co-immunoprecipitation analysis of protein lysates from non-transfected (Lane 1), HAP1/ARQ25-cotransfected (Lane 2), HAP1/ARQ65-cotransfected (Lane 3) and HAP1-transfected (Lane 4) HEp-2 cells. Each lysate was immunoprecipitated with a beads-labeled anti-AR antibody. Upper panels: western blot with an anti-AR antibody or anti-GFP antibody for samples before immunoprecipitation showing the presence of the approximately equal amounts of HAP1/ARQ25 or HAP1/ARQ65 in each cotransfected cell. Middle panel: western blot with an anti-AR antibody for the immunoprecipitated materials showing the presence of the approximately equal density of the two ARs. Lower panel: western blot with an anti-GFP antibody for the immunoprecipitated materials showing the polyQ-length-dependent interaction of AR with HAP1. Note that the GFP-immunopositive bands indicating HAP1 (asterisks) were more intensely stained in the pull-down of ARQ65 than that of ARQ25.
Effects of polyQ length on the HA-CAF ratio
In order to clarify the difference in the frequency or strength of association with HAP1 between ARQ25 and ARQ65, the HA-CAF ratios (HA-CAF cell number/total cotransfected cell number) were surrogated as an index and chronologically compared between HAP1/ARQ25- and HAP1/ARQ65-cotransfected cells. The HA-CAF ratios for ARQ25 and ARQ65 (HA25-CAF and HA65-CAF ratios) were 16% (81/506) and 22% (147/673) in 24 h, 31% (167/378: P <0.05) and 39% (159/410: P <0.05) in 48 h and 33% (110/327: P <0.05) and 44% (148/337: P <0.05) in 72 h after each cotransfection (Fig. 2 F). Thus, both the HA65-CAF and HA25-CAF ratios continued to increase during 72 h after the transfection, but the HA65-CAF ratio was 25–35% higher than the HA25-CAF ratio in any time lapse.
Co-immunoprecipitation of HAP1/ARQ25 and HAP1/ARQ65
Co-immunoprecipitation tests were performed in order to obtain biochemical evidence for the interaction of HAP1 with ARQ25 or ARQ65. Using protein lysates from GFP-HAP1/ARQ25- or /ARQ65-cotransfected HEp-2 cells, ARQ25 or ARQ65 was immunoprecipitated with a beads-labeled anti-AR antibody and boiled in order to dissociate proteins such as HAP1 and anti-AR antibody from the ARs. In the western blot with an anti-AR antibody or anti-GFP antibody for samples before immunoprecipitation, which were derived from the same ones as used in IP shown in the middle and lower panels of Figure 2 G, almost equal amounts of ARQ25 and ARQ65 were expressed between GFP-HAP1/ARQ25- and GFP-HAP1/ARQ65 cotransfected cells (Fig. 2 G: upper left panel), and almost equal amounts of GFP-HAP1 were also expressed between them (Fig. 2 G: upper right panel). In the western blot with an anti-AR antibody for the immunoprecipitated samples, bands with a similar intensity of immunostainings for ARQ25 and ARQ65 were obtained, indicating the presence of an approximately equal density of the two ARs in the resultant lysates (Fig. 2 G: middle panel). In addition, bands of GFP-HAP1 were also detected with an anti-GFP antibody at the expected molecular weight, indicating that HAP1 interacts with AR in HEp-2 cells. The bands were, however, more intensely immunostained in the pull-down of ARQ65 than that of ARQ25 (Fig. 2 G: lower panel).
Identification of HAP1-interacting domain in AR molecule
Cotransfection of cDNAs of HAP1 and AR-deletion mutants
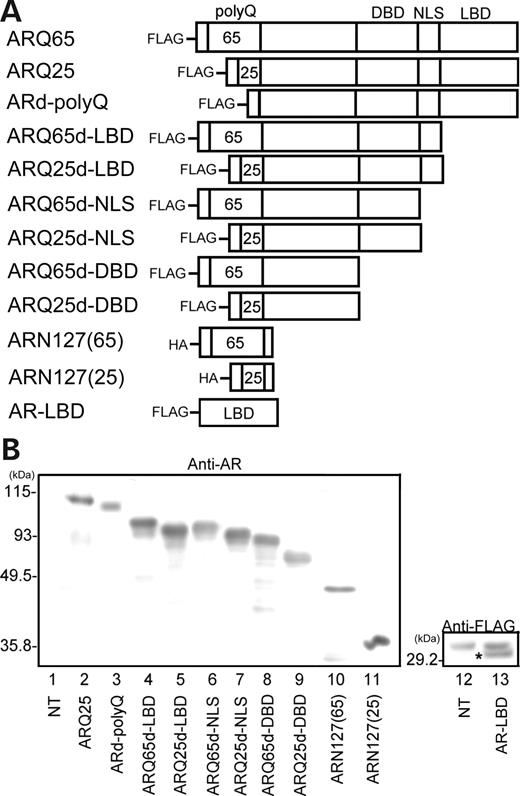
Several kinds of cDNAs of AR-deletion mutants shown in Figure 3 A were created in order to determine the region of AR crucial for interaction with HAP1. They include cDNAs encoding ARd-PolyQ (AR lacking the PolyQ tract), ARQ25/65d-LBD [ARQ25/65 lacking ligand-binding domain (LBD)], ARQ25/65d-NLS [ARQ25/65 lacking nuclear-location site (NLS) in addition to LBD], ARQ25/65d-DBD [ARQ25/65 lacking DNA-binding domain (DBD) in addition to NLS and LBD], and ARN127(65)/(25) [ARQ25/65 truncation mutant comprising common N-terminal 127-amino acids with 65 or 25 polyQ repeats]. In western blots with anti-AR and anti-FLAG-antibodies, cell lysates from the transfected HEp-2 cells revealed the expected molecular weight of these mutants (Fig. 3 B).
Construction of AR-deletion mutants. ( A ) Schematic representation of AR-deletion mutants of the ARQ25 and ARQ65. Each mutant is AR FLAG- or HA-tagged in the N-terminus. ARd-PolyQ, AR lacking the polyQ tract; ARQ25/65d-LBD, ARQ25/65 lacking LBD; ARQ25/65d-NLS, ARQ25/65 lacking NLS and LBD; ARQ25/65d-DBD, ARQ25/65 lacking DBD, NLS and LBD; ARN127(25)/(65), ARQ25/65 truncation-mutant comprising common N-terminal 127 amino acids with 65 or 25 polyQ repeats. ( B ) Western blots with anti-AR or anti-FLAG antibody for the lysates from cells transfected with each AR-deletion mutant, which confirms the expression of the corresponding AR-deletion mutants. Molecular weights (kDa) are indicated in the left of each panel. An asterisk indicates FLAG-tagged AR-LBD.
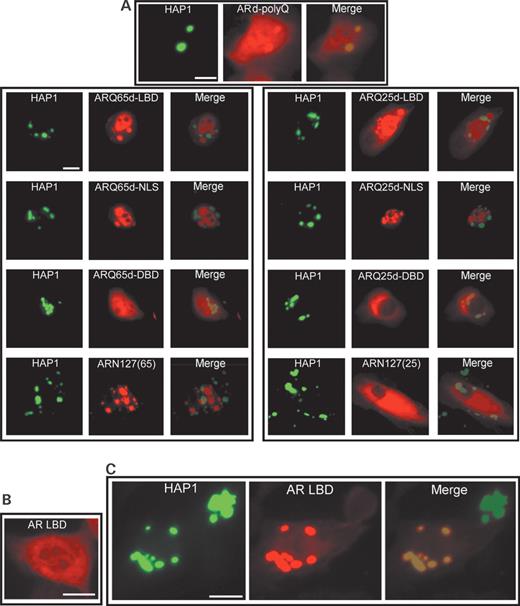
Cotransfection of cDNAs of HAP1 and ARd-PolyQ into HEp-2 cells produced HAP1/ARd-PolyQ-double-labeled cytoplasmic aggregations (Fig. 4 A: upper-most) but HA-CAF ratio of ARd-PolyQ is lower than those of ARQ25 and ARQ65 (data not shown). In contrast, all AR-deletion mutants lacking LBD [ARQ25/65d-LBD, ARQ25/65d-NLS, ARQ25/65d-DBD or ARN127(65)/(25)] did not form any HAP1/AR-deletion-mutant-double-labeled cytoplasmic aggregations (Fig. 4 A), indicating that the LBD region is essential for the association with HAP1 and that either the 25- or 65-polyQ repeat in AR itself is not a HAP1-interacting region.
Fluorescent photomicrographs showing subcellular interactions between HAP1 (GFP: green) and several kinds of AR-deletion mutants (Alexa594: red) in transfected HEp-2 cells. ( A ) Subcellular interactions between HAP1 and AR-deletion mutants without polyQ or LBD in their cotransfection. Note that ARd-polyQ without polyQ shows clear colocalization with HAP1 (uppermost), whereas no other merged image shows any colocalization of HAP1 and ARQ25- or ARQ65-deletion mutants without LBD (Fig. 3 ) (bar=10 µm). ( B ) Diffused subcellular distribution of AR-LBD in its single transfection (bar=10 µm). ( C ) Subcellular interactions between HAP1 and AR-LBD in their cotransfection. Note that AR-LBD alone can also interact with HAP1 as cytoplasmic inclusions (bar=10 µm).
Cotransfection of cDNAs of HAP1 and AR-LBD
The cDNA of FLAG-tagged LBD (AR-LBD) was made (Fig. 3 A) and transfected singly (Fig. 4 B) or cotransfected with HAP1 into HEp-2 cells (Fig. 4 C). Using an anti-FLAG antibody, the expression in the cultured cells was confirmed in western blotting (Fig. 3 B) and fluorescence microscopy (Fig. 4 B). In immunofluorescence cytochemistry, single transfections of the cDNA of AR-LBD always resulted in the diffuse expression of the products in the cytoplasm (less in the nucleus) (Fig. 4 B), but cotransfections with HAP1-cDNA usually induced the formation of prominent HAP1/AR-LBD-double-labeled cytoplasmic aggregations (Fig. 4 C). The results clearly show that the AR-LBD itself is an essential region for the association with HAP1, and that it is sequestered by the cytoplasmic HAP1-positive inclusions.
Effects of androgen on dynamism of AR and HAP1/AR complex
Effects of DHT on nuclear translocation of AR
The cDNA of ARQ25 or ARQ65 was transfected singly into HEp-2 cells in order to continuously observe changes of AR morphology. In control medium (DHT-undetectable medium before the addition of the androgen), most of the cells were observed to express ARQ25 or ARQ65 diffusely in the nuclei and cytoplasm in fluorescence microscopy (Fig. 5 A). Within 3 h after the addition of DHT into the transfected cells, both ARQ25 and ARQ65 were clearly translocated into the nuclei, making a sharp contrast with a prominently diminished cytoplasmic AR expression (Fig. 5 B and I). DHT effects on AR morphology were similar in ARQ25 and ARQ65 in HEp-2 cells.
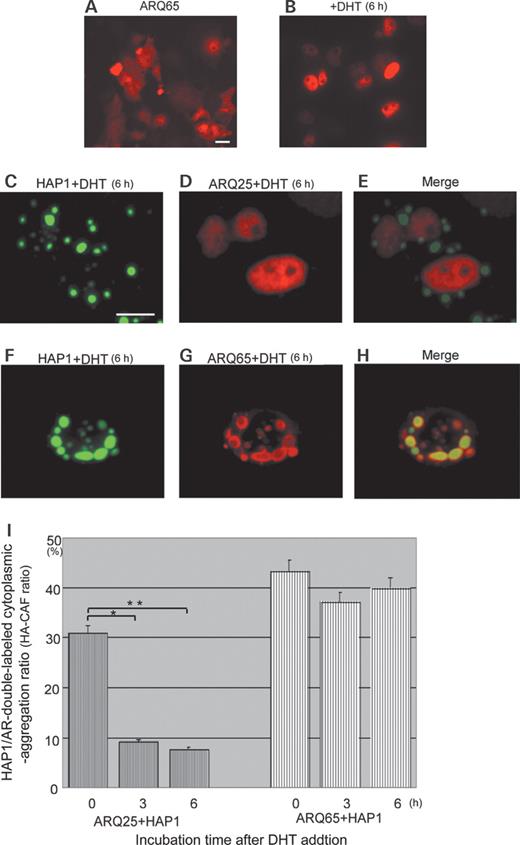
Effects of androgen on subcellular interaction between HAP1 (GFP: green) and ARQ25 or ARQ65 (Alexa594: red) in the cotransfected HEp-2 cells ( A and B ) Fluorescent photomicrographs showing that diffused cytoplasmic ARQ65 is translocated into the nucleus 6 h after DHT administration (10 −6 M ) (bar=10 µm). ( C – E ) Fluorescent photomicrographs showing that the HAP1/ARQ25 complex is dissociated 6 h after DHT administration (10 −6 M ). Note that ARQ25 is translocated into the nucleus, whereas HAP1 remains as a cytoplasmic inclusion (bar=10 µm). ( F – H ) Fluorescent photomicrographs showing persistence of the HAP1/ARQ65 complex even 6 h after DHT administration. Note that HAP1 and ARQ65 stay colocalized in most of the cytoplasmic inclusions (bar=10 µm). ( I ) Bar graph comparing HA-CAF ratios in time course between ARQ25 and ARQ65 (0, 3 and 6 h) after DHT administration (10 −6 M ). Note that the HA-CAF ratio after DHT administration is kept stable in the case of ARQ65+HAP1 when compared with that of ARQ25+HAP1 showing the prominent decrease in time course. Asterisks indicate statistical difference in HA-CAF ratios of ARQ25 between 0 and 3 h ( P <0.05) and between 0 and 6 h.
Effects of DHT on HA25- and HA65-CAF ratios
In order to examine the effects of DHT on the association of ARQ65 or ARQ25 with HAP1, cDNAs of GFP-HAP1 and ARQ65 or ARQ25 were cotransfected into HEp-2 cells. The HA65-CAF and HA25-CAF ratios were compared over time after DHT administration (0, 3 and 6 h). In the case of ARQ25, addition of DHT induced nuclear translocation of ARQ25 independently of with or without HAP1 inclusions (Fig. 5 C, D and E). The HA25-CAF ratio at 0 h (pre-administration control) was 31% (117/378, P <0.05). The ratio was dramatically decreased to 9% (19/207, P <0.05) in 3 h and 7% (15/197, P <0.05) in 6 h after DHT administration (Fig. 5 I). In contrast, in the cotransfection of ARQ65 with HAP1, the double-labeled cytoplasmic inclusions showed little change, with DHT being high throughout the experiment. Compared with ARQ25, the nuclei showed much weaker ARQ65 expression marked by little or no Alexa594-fluorescence (Fig. 5 F, G and H). The HA65-CAF ratio was 43% (124/286) at 0 h, 37% (178/478) in 3 h and 40% (151/378) in 6 h after DHT administration (Fig. 5 I). The results indicate that the addition of DHT specifically reduces the association of HAP1 with ARQ25, but does not clearly affect that of HAP1 with SBMA-mutant ARQ65 in 6 h.
Effects of androgen and HAP1-cotransfection on ARQ65-induced apoptosis
Effects of DHT on apoptosis in ARQ25- or ARQ65-transfected cells
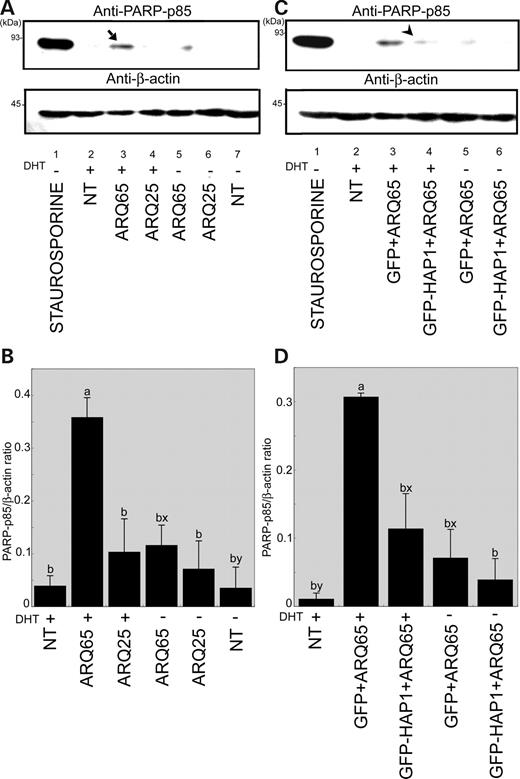
In order to examine the cellular toxicity of DHT via AR, the induction of intracellular poly-ADP-ribose polymerase p85 fragment (PARP-p85), a caspase-3-cleaved form regarded as a specific marker for apoptosis, was analyzed by western blot with an anti-PARP-p85 antibody in lysates from ARQ25- or ARQ65-transfected cells cultured in a non-DHT control or DHT-added medium (Fig. 6 A and B). Each integrated density value (IDV) for PARP-p85 expression was normalized to that for β-actin expression as a PARP-p85-IDV/β-actin-IDV (PARP-p85/β-actin) ratio. The quantitative data, which were finally presented as the averaged ratios (mean±SEM), revealed that the PARP-p85/β-actin ratio was much higher in the DHT-added ARQ65-transfected cells than in DHT-added ARQ25-transfected cells (Fig. 6 B: 3.48-fold, P <0.01). In non-DHT added ARQ65-transfected cells, the PARP-p85/β-actin ratio was much lower than in DHT-added ARQ65-transfected cells (Fig. 6 B: 0.32-fold, P <0.01). The PARP-p85/β-actin ratios in non-DHT-added NT and ARQ25-transfected cells showed no significant difference when compared with those in DHT-added NT or ARQ25-transfected cells. The results show that cellular apoptosis is induced by DHT, and that it occurs more specifically and strongly in ARQ65-transfected cells than in ARQ25-transfected cells.
Effects of androgen and HAP1-cotransfection on ARQ65-induced apoptosis. ( A ) Western blot analysis with a anti-PARP-p85 antibody for apoptosis comparing among lysates from non-, ARQ25- and ARQ65-transfected cells cultured under a control (−) or DHT-added (+) (6 h) condition (upper panel). Lane 1, staurosporine-treated non-transfected cells in control medium (positive control); Lane 2, DHT-treated non-transfected cells; Lane 3, DHT-treated ARQ65-transfected cells; Lane 4, DHT-treated ARQ25-transfected cells; Lane 5, ARQ65-transfected cells in control medium; Lane 6, ARQ25-transfected cells in control medium; Lane 7, non-transfected cells in control medium. Western blot for β-actin was used as a loading control (lower panel). Note that apoptosis is prominently induced in DHT-treated ARQ65-transfected cells (an arrow in Lane 3). Molecular weights (kDa) are indicated in the left of each panel. ( B ) Quantification of the PARP-p85 normalized to β-actin. Each data is presented as mean±SEM of PARP-p85/β-actin ratio obtained from three independent experiments (a and b: P <0.01 for PARP-p85; x and y: P <0.05 for PARP-p85). ( C ) Western blot analysis with an anti-PARP-p85 antibody for apoptosis comparing among lysates from non-transfected, GFP/ARQ65-cotransfected or GFP-HAP1/ARQ65-cotransfected cells cultured under a control (−) or DHT-added (+) (6 h) condition. Lane 1, staurosporine-treated non-transfected cells in control medium (positive control); Lane 2, DHT-treated non-transfected cells; Lane 3, DHT-treated GFP/ARQ65-cotransfected cells; Lane 4, DHT-treated GFP-HAP1/ARQ65-cotransfected cells; Lane 5, GFP/ARQ65-transfected cells in control medium; Lane 6, GFP-HAP1/ARQ65 cotransfected cells in control medium. Western blot for β-actin was used as a loading control (lower panel). Note that the DHT-induced apoptosis seen in GFP/ARQ65-cotransfected cells (Lane 3) is prominently suppressed in GFP-HAP1/ARQ65-cotransfected cells (an arrowhead in Lane 4) as well as non-DHT-treated GFP/ARQ65-cotransfected cells (Lane 5). Molecular weights (kDa) are indicated in the left of each panel. ( D ) Quantification of the PARP-p85 normalized to β-actin. Each data is presented as mean±SEM of PARP-p85/β-actin ratios obtained from three independent experiments (a and b: P <0.01 for PARP-p85; x and y: P <0.05 for PARP-p85).
Effects of HAP1-cotransfection on DHT-induced apoptosis in ARQ65-transfected cells
In order to examine the effects of HAP1-cotransfection on DHT-induced apoptosis in ARQ65-transfected cells, PARP-p85 expression detected by western blot with an anti-PARP-p85 antibody was compared among HEp-2 cells, which were cotransfected with ARQ65/GFP or ARQ65/GFP-HAP1 and cultured under the condition with or without DHT (Fig. 6 C and D). The quantitative data revealed that the highest PARP-p85/β-actin ratio was obtained in the DHT-added GFP/ARQ65-cotransfected cells (DHT-added GFP/ARQ65 control). On the other hand, the PARP-p85/β-actin ratio in DHT-added GFP-HAP1/ARQ65-cotransfected cells was much lower than that seen in the aforementioned control (Fig. 6 D; 0.36-fold, P <0.01), and showed no significant difference when compared with that in non-DHT-added GFP/ARQ65 cotransfected cells (non-DHT-GFP/ARQ65 control). These results indicate that HAP1 cotransfection suppresses DHT-induced apoptosis in ARQ65-transfected cells as significantly as DHT deprivation.
DISCUSSION
HAP1 interacts with AR via LBD in a polyQ-dependent manner and forms cytoplasmic aggregations
The present in vitro study, employing western blot, immunoprecipitation and intracellular transfection of several kinds of AR mutant and GFP-fused-HAP1 cDNAs, provides the first striking evidence that HAP1 interacts with AR in an AR-polyQ-length-dependent manner and forms prominent cytoplasmic aggregations sequestering AR. Deletion of LBD strongly inhibits the interaction of HAP1 with AR. Intracellular cotransfection of HAP1 and AR-LBD results in the formation of the prominent cytoplasmic aggregation of the HAP1/AR-LBD complex. The data indicate that LBD is an essential region where HAP1 can interact with AR. Thus, HAP1 could affect LBD and modify the AR functions elicited by androgens. It is yet to be determined, however, whether HAP1 can bind to AR-LBD directly or indirectly. To address this question, further analyses (i.e. yeast two-hybrid assay, in vitro binding assay, and so on) are required.
As a HA-CAF ratio is augmented in a polyQ-length-dependent manner with or without DHT, the expansion of polyQ may affect the three-dimensional structure of AR and enhance the accessibility of HAP1 to the AR-LBD. A normal length of polyQ-repeat motif might be prone to inhibiting an approach of HAP1 to the LBD to some extent and regulate the interaction. The longer expansion of polyQ might expose AR-LBD and enable HAP1 to access it more readily.
Both HA65-CAF and HA25-CAF ratios keep increasing after their transfection, but the former ratio is 30% higher than the latter, indicating that the association of HAP1 with ARQ65 is stronger than that with ARQ25, independent of time after HAP1- and AR-cotransfection. In the presence of DHT, the HA25-CAF ratio was dramatically decreased to less than the one-third of the CAF ratio in the non-DHT control. In contrast, the HA65-CAF ratio was affected by less than 10% even 6 h after the addition of DHT. Thus, in the presence of sufficient androgen, the HA65-CAF ratio was 5–6-fold higher than the HA25-CAF ratio. Addition of DHT appears to dissociate the HAP1/ARQ25 complexes, whereas most of the HAP1/ARQ65 complexes remain as stable aggregations even at increased androgen levels. As the DHT-binding to AR would subsequently cause phosphorylation and other conformational changes in the AR morphology, it is speculated that the conformational changes might reduce the association strength of HAP1 with ARQ25, but not substantially affect that with ARQ65. Alternatively, as HAP1 interacts with AR-LBD, HAP1 might compete with androgens for AR-LBD. In either case, the much stronger association of ARQ65 with HAP1 suggests that HAP1/AR complexes could be more frequently formed in SBMA disease. Some of these could grow into distinct inclusions in androgen-sensitive regions of the patients. Cytoplasmic inclusions are often seen in otherwise unchanged brain tissue of SBMA patients ( 29 ).
It is also of importance to note that HAP1 can interact with ARQ25 to some extent, even in the presence of androgens. HAP1 might modify masculinization or defeminization of the brain via polyQ-length-dependent regulation of neural AR functions (Fig. 7 A and B).
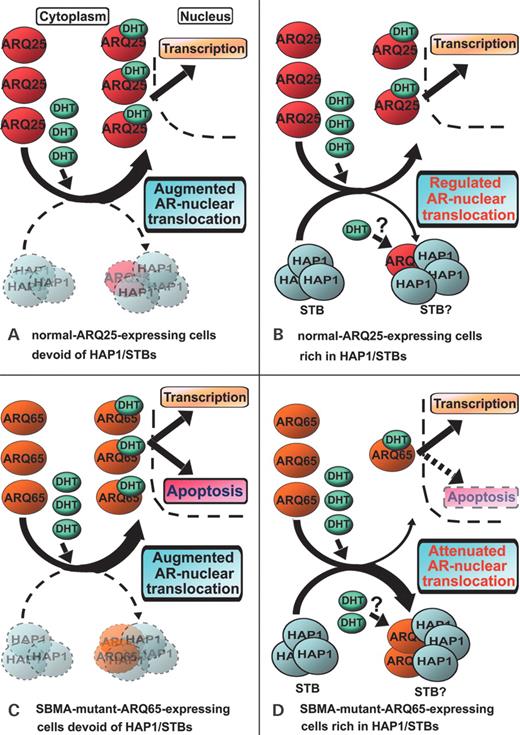
Models for ‘subcellular regulation or attenuation of AR-nuclear translocation by HAP1/STBs’. ( A ) When DHT is increased in normal ARQ25-expressing cells devoid of HAP1/STBs, AR-nuclear translocation is augmented, leading to enhanced gene transcription in the down stream of AR-responsive element (ARE). ( B ) When DHT is increased in normal ARQ25-expressing cells rich in HAP1/STBs, AR-nuclear translocation is regulated by sequestration into cytoplasmic HAP1/STBs to some extent, which might control gene transcription in the downstream of ARE. ( C ) When DHT is increased in SBMA-mutant-ARQ65-expressing cells devoid of HAP1/STBs, AR-nuclear translocation is augmented, inducing apoptosis probably by enhanced ARQ65 truncation and/or intranuclear accumulation of the toxic fragments; androgen functions might be exacerbated by ARQ65-expressing cell dysfunction. ( D ) When DHT is increased in SBMA-mutant-ARQ65-expressing cells rich in HAP1/STBs, AR-nuclear translocation is attenuated and regulated by sequestration into cytoplasmic HAP1/STBs, suppressing SBMA-mutant-ARQ65-induced apoptosis, probably by blocking ARQ65 truncation and/or reducing the intranuclear accumulation of the toxic fragments; androgen functions might be relatively brought forward by the recovery of ARQ65-expressing cell functions. This model might explain that our previous ‘HAP1 protection hypothesis’ in HD can also be applied to SBMA.
HAP1 expression protects DHT-induced apoptosis particularly in ARQ65-transfected cells
It has been previously shown that expanded-polyQ-AR in transfected cells is toxic and leads to apoptosis ( 30 , 31 ). Our present western blot analysis for PARP-p85 demonstrated that the transfection of ARQ65 induces prominent apoptosis in the presence of DHT (Fig. 6 A and B).
The most exciting result in the present study was that transfected HAP1 expression clearly suppresses the ARQ65-induced apoptosis in vitro as effectively as androgen deprivation (Fig. 6 C and D). The cell toxicity of expanded-polyQ-AR has been argued to be primarily attributable to nuclear translocation of its truncated N-terminus, though it remains to be determined whether a truncation of the N-terminus occurs in the cytoplasm or nucleus ( 25 , 32 , 33 ). Bindings of DHT or other androgens to AR strongly induce the nuclear translocation of AR, probably via a nuclear localization signal (NLS) domain near AR-LBD, which is disclosed by AR-conformational changes after the binding of androgens ( 34–36 ). Putative caspase-3-acting sites enhancing N-terminus truncation of AR might become more accessible in longer-polyQ expanded AR, as is inferred in polyQ-Htt ( 9 , 37 , 38 ). Like other molecular chaperones (HSP90 and HSP70), HAP1, which is much larger in molecular size than androgens, might not only interact with the LBD but also affect or cover the adjacent NLS and putative neighboring caspase-3-acting sites, preventing ARQ65 from being translocated into the nucleus prior to truncation into toxic fragments. This is in sharp contrast to the action of androgens promoting the nuclear translocation and truncation of ARQ65 by putative disclosure of NLS and caspase-3-acting sites in AR. Alternatively, the putative suppression of nuclear translocation might be due to size-dependent inhibition related to the formation of HAP1/AR inclusions. HAP1/AR complexes were prone to assembling into solitary cytoplasmic inclusions or huge aggregation clusters surrounding the nucleus. Transfection of HAP1cDNA has been reported to induce the STBs as solitary spherical HAP1 inclusions ( 7 ). AR tended to localize in the periphery of the aggregation complexes and, in particular, show a ring-shaped appearance in the solitary inclusions (Figs. 1 and 5 ). Thus most of the inclusions observed in the present in vitro study are inferred to be the STBs sequestering AR, whereas some aggregations might be clusters of small HAP1/AR molecules without growing into the typical STB. Detailed electron microscopic analysis is required for more precise identification of those structures. In either case, both of them appear to be too large to be translocated into the nucleus. The NLS domain might be concealed or embedded inside the packed conglomeration. Recent studies indicate that the retention of perikaryal AR is not consistently cytotoxic. The cytoplasmic accumulation may indicate resistance to AR nuclear translocation with increased likelihood of cell-survival ( 32 ). Abnormally expanded-polyQ-AR could be more strongly sequestered by HAP1 inclusion than normal length polyQ-AR. This suggests that intracellular HAP1 induction might prevent its toxic actions more efficiently in SBMA patients.
Other possible suppressive mechanisms might indirectly involve the activation of other protective pathways through the regulation of other HAP1-binding partners, such as NeuroD (ND) and/or epidermal growth factor (EGF) receptor (EGFR) ( 39 ). The overexpression of ND results in premature neuronal differentiation and ectopic neurogenesis in Xenopus embryos ( 40 ), whereas the disruption of ND in mice causes massive cell death in subsets of differentiating and mature neurons ( 41–43 ) and induces the transcriptional downregulation of several genes, such as a gene for brain-derived neurotrophic factor (BDNF) ( 44–46 ). Studies have shown that deletion of EGFR results in postnatal neurodegeneration and death in mice ( 47 , 48 ). Overexpressed HAP1 protects against EGFR degradation through another HAP1 interactor, the human hepatocyte-related tyrosine kinase substrate (Hrs), which might be involved in the endosome-to-lysosome trafficking of EGFR ( 49 ). Sheng et al . ( 50 ) have recently reported that a fasting stress increases the hypothalamic HAP1 expression, which would in turn facilitate a feeding behavior via its upregulation of the γ-aminobutyric acid type A (GABA A ) receptor. In contrast, Hap1-KO mice have been demonstrated to be vulnerable to the fasting stress because of a depressed feeding behavior owing to increased neuronal apoptosis in the fetal and neonatal hypothalamus ( 5 , 6 ). HAP1 might play an important role in protecting neurons from several kinds of cellular stress in postnatal premature animals. Disruption of the putative protective pathways is thus inferred to enhance the vulnerability to neuronal stress and to insult self- and/or species-preservation systems in the hypothalamic and limbic regions ( 9 ), which would result in early postnatal death as occurred in the KO mice for HAP1 or the aforementioned HAP1-associated proteins.
Possible involvement of HAP1 in SBMA pathogenesis: therapeutic possibility
Expanded polyQ-AR is a pathogenetic protein for SBMA, an adult-onset X-linked spinal and bulbar motor neurodegenerative disorder characterized by progressive proximal (bulbar and limb) muscle atrophy, weakness and fasciculations ( 22 , 26 , 27 , 51 ). SBMA also involves subclinical mild sensory impairment ( 24 , 26 , 52 , 53 ). Elevated serum creatine kinase concentrations, impaired glucose tolerance, hepatic dysfunction and hyperlipidemia, suggest visceral involvement ( 26 , 52 ). The mutant AR gene has up to 68 repeats, whereas the normal number of CAG repeats is 11–36. An inverse correlation exists between the CAG repeat size and age at onset, as well as disease severity in SBMA ( 23 , 54 , 55 ). Prominent SBMA symptoms occur exclusively in men, being frequently accompanied by androgen insensitivity with gynecomastia, testicular atrophy and reduced fertility ( 56 , 57 ). As severe testicular feminization patients in whom AR is fully inactivated, do not have motor neuron disease ( 58 , 59 ), motor dysfunctions in SBMA has been construed to be attributable to abnormal function of the N-terminus truncated mutant AR, accompanied by its hormone-dependent nuclear translocation ( 25 , 32 ). Castration or treatment with an LHRH agonist (i.e. leuprolerin) reducing testosterone levels has recently been reported to dramatically improve the impaired survival rate and neuromuscular dysfunction seen in transgenic mice carrying the full-length human AR with expanded polyQ tracts, whereas symptoms were exacerbated by administration of testosterone or an androgen antagonist (i.e. flutamide) promoting nuclear translocation ( 60 ). SBMA appears to involve two different, but closely interrelated aspects of AR dysfunction, ‘a loss of normal AR function’ in masculinization or defeminization and ‘a gain of abnormal AR function’ in the bulbar and spinal muscular system.
The recent in situ hybridization study ( 9 ) indicated that HAP1 is preferentially localized to the limbic and hypothalamic regions, where neurodegeneration rarely occurs in HD and SBMA, whereas the target lesions, including striatum, thalamus, neocortex and lower brainstem (bulbar regions), show much weaker HAP1 expression. A conundrum in SBMA pathogenesis concerns why most of the forebrain regions rich in AR are spared from neurodegeneration unlike similarly AR-expressive bulbar and spinal motor neurons. Neurons with AR or ERα in the hypothalamus and limbic regions have been pointed out to coexpress HAP1/STBs at a high frequency ( 1 , 9 , 20 ). In fact, a large number of neurons in the rodent forebrain coexpress both AR and HAP1 (our unpublished data), though sufficient information has not yet been obtained whether or not AR is localized to the STB in such neurons under a normal androgen condition. In contrast, most bulbar motor neurons (e.g. trigeminal motor nucleus, facial nucleus, glossopharyngeal nucleus, hypoglossal nucleus) and spinal motor neurons in the anterior horn have been reported to express AR ( 61 , 62 ), but not so much HAP1 ( 9 ). It has been reported that in vitro transfection of HAP1 partially inhibit the activation of caspase-3 and suppresses apoptosis in Htt-polyQ-transfected HEK293 cells ( 6 ). The present in vitro results indicate that SBMA-mutant-AR-induced apoptosis is more selectively suppressed in neurons with HAP1/STB than in those without HAP1/STB, suggesting that the ‘HAP1 protection hypothesis’ proposed in HD ( 9 ) might be extended to SBMA pathogenesis. Thus, the region-specific coexpression of HAP1 and AR may account for the region-specific protection against neurodegeneration in SBMA.
Considering that SBMA female and young male patients are almost intact, most of SBMA-mutant AR is thought to function normally and only the minor part might be truncated into a toxic form and translocated into the nucleus by androgen. Neurodegeneration (or apoptosis) underlying manifestation might not occur until the nuclear accumulation of the toxic form is driven by androgen and exceeds over a certain threshold. If the nuclear translocation of AR could be restricted therapeutically, its toxic aspects of the SBMA-mutant AR might be attenuated while permitting some degree of normal function to be expressed (Fig. 7 ). Intracellular HAP1 induction might not only suppress apoptosis by cytoplasmic sequestration of SBMA-mutant AR, but also permit some normal androgenic activity via the mutant AR. Androgen actions other than via nuclear AR are thought to stay intact in SBMA. They may include those through the androgen's allosteric modification of several neurotransmitter receptors ( 63 ) and activation of a distinct membrane AR as recently suggested in lower vertebrates ( 64 , 65 ). Unlike androgen-deprivation, the HAP1 induction would not completely block androgen actions. Its therapeutic applications might prevent neurodegeneration and ameliorate clinical dysfunction in SBMA while avoiding the effects of androgen deprivation by surgical or chemical castration (e.g. impotence, muscle weakness and lowered drive and motivation) ( 25 ).
The present study provides clear evidence that HAP1 can interact with subcellular AR via LBD to attenuate or regulate AR functions and that the interaction is enhanced in an AR-polyQ-length-dependent manner to suppress the toxic effects. These results suggest that HAP1/STB acts in SBMA as it does in HD as an important intrinsic neuroprotectant. HAP1 is expressed abundantly and preferentially in the limbic and hypothalamic regions, which are relatively spared from neurodegeneration in HD and SBMA. Its low expression in affected regions provides a likely explanation from the distribution of neurodegeneration in HD and SBMA. The level of expression of HAP1/STB might determine the threshold for vulnerability of the cell to apoptosis in HD and SBMA. It would be of great interest to investigate if neuroprotective HAP1/STB levels are more widely significant in the pathogenesis of several other neurodegenerative diseases, particularly those with triplet-repeats, including dentatorubral-pallidoluysian atrophy, spinocerebellar ataxias, fragile X syndrome, myotonic dystrophy and Friedreich ataxia.
MATERIALS AND METHODS
Construction of full-length and mutant AR plasmids
The HAP1 cDNA obtained from rat brains by RT–PCR ( 9 ) were cloned into the Xho I– Eco RI-digested pEGFP-C2 vector (Clontech) for the construction of N-terminus GFP-fused-HAP1-cDNA plasmid; HAP1 designated here corresponds to HAP1A, the most common type of HAP1, which exists in the human. The cDNAs encoding hemagglutinin (HA)-tagged human ARQ25 and ARQ65 ( 28 ) were generously gifted from Dr Keith R. Yamamoto (School of Medicine, University of California, San Francisco, USA) and utilized to re-construct FLAG-tagged full-length ARQ25 and ARQ65 using hARcN2 and hARcC2 primers, which were then inserted into the Bgl II– Bam HI-digested p3×FLAG-CMV-10 vector (Sigma). The amino acid sequences of AR were on the basis of the study of ARQ22 ( 66 ) and standardized to ARQ25 in the present study. The ARd-PolyQ (lacking amino acids 58–82 in ARQ25) was prepared by ligation of the 5′-fragment-cDNA encoding 1–57 amino acids of AR and 3′-fragment encoding 84–921 amino acids at the Xba I site; the former were amplified by PCR with hARcN2 and hARc-d-QAs primers, whereas the latter were amplified with hARc-d-Qs and hARcC2 primers. The resultant fragment, which was amplified by PCR using hARcN2 and hARC2 primers, was inserted into the Bgl II– Bam HI-digested p3×FLAG-CMV-10 vector. On the basis of a previous report concerning the domain structure of AR ( 66 ), the other series of cDNAs for several AR domain-specific-deleted mutants, including ARd-LBD (lacking amino acids 670–921 in ARQ25), ARd-NLS (lacking amino acids 622–921 in ARQ25), ARd-DBD (lacking amino acids 529–921 in ARQ25) and AR-LBD (amino acids 669–921 in ARQ25), were amplified from the cDNA encoding ARQ25 or ARQ65 by PCR with the following primers (ARd-LBD; hARcN1 and hARc-d-LBD, ARd-NLS; hARcN1 and hARc-d-NLS, ARd-DBD; hARcN1 and hARc-d-DBD, AR-LBD; hARc-LBD and hAR-cC2). These PCR products were purified and ligated into the Eco RI– Bgl II-digested (for AR-d-LBD, AR-d-NLS, AR-d-DBD) or Bgl II– Bam HI-digested (for AR-LBD) p3×FLAG-CMV-10 vector. ARN127(25) and ARN127(65) were also gifted from Dr Keith R. Yamamoto and used as it is ( 28 ). The primer sequences and plasmids for AR and AR mutants used in this study are listed in Table 1 .
Primers used for the construction of expression plasmids for AR mutants
| Primers . | Sequences a . | Restriction enzymes . | Used to construct . |
|---|---|---|---|
| hARcN1 | 5′-GG GAATTC GATGGAAGTGCAGTTAGGGCTG-3′ | Eco RI | ARQ65d-LBD, ARQ65d-NLS, ARQ65d-DBD, ARQ25d-LBD, ARQ25d-NLS, ARQ25d-DBD |
| hARc-d-LBD | 5′-TCC AGATCT TAGCCTTCAATGTGTGACAC-3′ | Bgl II | ARQ65d-LBD, ARQ25d-LBD |
| hARc-d-NLS | 5′-CC AGATCT CATTTCCGAAGACGACAAGATG-3′ | Bgl II | ARQ65d-NLS, ARQ25d-NLS |
| hARc-d-DBD | 5′-TCC AGATCT CAGGGGCCCATTTCGCTTTTG-3′ | Bgl II | ARQ65d-DBD, ARQ25d-DBD |
| hARcLBD | 5′-CG AGATCT GGAATGTCAGCCCATCTTTCTG-3′ | Bgl II | AR-LBD |
| hARcN2 | 5′-GA AGATCT GATGGAAGTGCAGTTAGGGCTG-3′ | Bgl II | ARQ65, ARQ25, ARd-polyQ |
| hARcC2 | 5′-TGC GGATCC TCACTGGGTGTGGAAATAG-3′ | Bam HI | AR-LBD, ARQ65, ARQ25, ARd-polyQ |
| hARc-d-QAs | 5′-TAA TCTAGA CAGCAGCAGCAAACTGGCGCC-3′ | Xba I | ARd-polyQ |
| hARc-d-Qs | 5′-ATT TCTAGA GAGACTAGCCCCAGGCAGACG-3′ | Xba I | ARd-polyQ |
| Primers . | Sequences a . | Restriction enzymes . | Used to construct . |
|---|---|---|---|
| hARcN1 | 5′-GG GAATTC GATGGAAGTGCAGTTAGGGCTG-3′ | Eco RI | ARQ65d-LBD, ARQ65d-NLS, ARQ65d-DBD, ARQ25d-LBD, ARQ25d-NLS, ARQ25d-DBD |
| hARc-d-LBD | 5′-TCC AGATCT TAGCCTTCAATGTGTGACAC-3′ | Bgl II | ARQ65d-LBD, ARQ25d-LBD |
| hARc-d-NLS | 5′-CC AGATCT CATTTCCGAAGACGACAAGATG-3′ | Bgl II | ARQ65d-NLS, ARQ25d-NLS |
| hARc-d-DBD | 5′-TCC AGATCT CAGGGGCCCATTTCGCTTTTG-3′ | Bgl II | ARQ65d-DBD, ARQ25d-DBD |
| hARcLBD | 5′-CG AGATCT GGAATGTCAGCCCATCTTTCTG-3′ | Bgl II | AR-LBD |
| hARcN2 | 5′-GA AGATCT GATGGAAGTGCAGTTAGGGCTG-3′ | Bgl II | ARQ65, ARQ25, ARd-polyQ |
| hARcC2 | 5′-TGC GGATCC TCACTGGGTGTGGAAATAG-3′ | Bam HI | AR-LBD, ARQ65, ARQ25, ARd-polyQ |
| hARc-d-QAs | 5′-TAA TCTAGA CAGCAGCAGCAAACTGGCGCC-3′ | Xba I | ARd-polyQ |
| hARc-d-Qs | 5′-ATT TCTAGA GAGACTAGCCCCAGGCAGACG-3′ | Xba I | ARd-polyQ |
a Italicized sequences indicate restriction enzyme sites.
Primers used for the construction of expression plasmids for AR mutants
| Primers . | Sequences a . | Restriction enzymes . | Used to construct . |
|---|---|---|---|
| hARcN1 | 5′-GG GAATTC GATGGAAGTGCAGTTAGGGCTG-3′ | Eco RI | ARQ65d-LBD, ARQ65d-NLS, ARQ65d-DBD, ARQ25d-LBD, ARQ25d-NLS, ARQ25d-DBD |
| hARc-d-LBD | 5′-TCC AGATCT TAGCCTTCAATGTGTGACAC-3′ | Bgl II | ARQ65d-LBD, ARQ25d-LBD |
| hARc-d-NLS | 5′-CC AGATCT CATTTCCGAAGACGACAAGATG-3′ | Bgl II | ARQ65d-NLS, ARQ25d-NLS |
| hARc-d-DBD | 5′-TCC AGATCT CAGGGGCCCATTTCGCTTTTG-3′ | Bgl II | ARQ65d-DBD, ARQ25d-DBD |
| hARcLBD | 5′-CG AGATCT GGAATGTCAGCCCATCTTTCTG-3′ | Bgl II | AR-LBD |
| hARcN2 | 5′-GA AGATCT GATGGAAGTGCAGTTAGGGCTG-3′ | Bgl II | ARQ65, ARQ25, ARd-polyQ |
| hARcC2 | 5′-TGC GGATCC TCACTGGGTGTGGAAATAG-3′ | Bam HI | AR-LBD, ARQ65, ARQ25, ARd-polyQ |
| hARc-d-QAs | 5′-TAA TCTAGA CAGCAGCAGCAAACTGGCGCC-3′ | Xba I | ARd-polyQ |
| hARc-d-Qs | 5′-ATT TCTAGA GAGACTAGCCCCAGGCAGACG-3′ | Xba I | ARd-polyQ |
| Primers . | Sequences a . | Restriction enzymes . | Used to construct . |
|---|---|---|---|
| hARcN1 | 5′-GG GAATTC GATGGAAGTGCAGTTAGGGCTG-3′ | Eco RI | ARQ65d-LBD, ARQ65d-NLS, ARQ65d-DBD, ARQ25d-LBD, ARQ25d-NLS, ARQ25d-DBD |
| hARc-d-LBD | 5′-TCC AGATCT TAGCCTTCAATGTGTGACAC-3′ | Bgl II | ARQ65d-LBD, ARQ25d-LBD |
| hARc-d-NLS | 5′-CC AGATCT CATTTCCGAAGACGACAAGATG-3′ | Bgl II | ARQ65d-NLS, ARQ25d-NLS |
| hARc-d-DBD | 5′-TCC AGATCT CAGGGGCCCATTTCGCTTTTG-3′ | Bgl II | ARQ65d-DBD, ARQ25d-DBD |
| hARcLBD | 5′-CG AGATCT GGAATGTCAGCCCATCTTTCTG-3′ | Bgl II | AR-LBD |
| hARcN2 | 5′-GA AGATCT GATGGAAGTGCAGTTAGGGCTG-3′ | Bgl II | ARQ65, ARQ25, ARd-polyQ |
| hARcC2 | 5′-TGC GGATCC TCACTGGGTGTGGAAATAG-3′ | Bam HI | AR-LBD, ARQ65, ARQ25, ARd-polyQ |
| hARc-d-QAs | 5′-TAA TCTAGA CAGCAGCAGCAAACTGGCGCC-3′ | Xba I | ARd-polyQ |
| hARc-d-Qs | 5′-ATT TCTAGA GAGACTAGCCCCAGGCAGACG-3′ | Xba I | ARd-polyQ |
a Italicized sequences indicate restriction enzyme sites.
Cell culture and transfection
The human larynx carcinoma cell line HEp-2 was routinely maintained in Dulbecco's-modified Eagle's-medium (DMEM) (Gibco BRL) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS) and antibiotics (penicillin G sodium, 100 UI/ml and streptomycin sulfate 100 µg/ml) and grown at 37°C in humidified atmosphere (5% CO 2 –95% air). Cytotoxicity with full-length polyQ-expanded AR is not always successfully obtained in cultured cells, except those in some sensitized treatment ( 67 ). To our experience, however, that depends on cell-lines; COS-7, HEK293 and HeLa cells appear to be inappropriate for the detection of cytotoxicity for full-length polyQ-expanded AR, whereas HEp-2 cells were more suitable for the current assessment. Endogenous HAP1 was not detected in non-transfected HEp-2 cells by western blotting or immunocytochemistry with four kinds of anti-HAP1 antibodies [HAP1N18; D19; C19, Santa Cruz Biotechnology; HAP1R12, recently prepared in our laboratory (unpublished)] (data not shown). Androgen concentrations (testosterone and DHT) in the culture medium used in the present study were measured by enzyme immunoassay using ELISA kits (IBL-Hamburg GmBH) and were at under-detection levels; less than sensitivities for testosterone (83 pg/ml) and for DHT (6 pg/ml). The morphology of exogenously expressed AR in HEp-2 cells in this culture medium was similar to that in phenol-red-free DMEM supplemented with 10% charcoal-treated FBS. Thus, the DMEM supplemented with the same batch of the FBS was used as a ‘control medium’ in the current study. Jet PEI transfection reagent (Polyplus-Transfection) was used for the transient transfection, according to the manufacturer's instructions.
Western blot and data analyses
After the transfection of several cDNAs, or the addition of DHT (5α-androstan-17β-ol-3-one, Fluka Chemie AG) or staurosporine (Sigma–Aldrich), the cells were collected, lysed, sonicated and boiled in sodium-dodecyl-sulfate (SDS) sample buffer. The extracted proteins were separated using SDS/polyacrylamide-gel electrophoresis (SDS/PAGE) and transferred onto PVDF membrane. The membrane was blocked with 5% skim-milk in Tris-buffered saline (20 m M Tris–HCl, 137 m M NaCl; pH 7.6) containing 0.1% Tween-20 (TBST) for 1 h at 20°C and incubated overnight at 4°C with primary antibodies diluted 1:1000 [a rabbit anti-AR polyclonal antibody (ARN-20, Santa Cruz Biotechnology), a rabbit anti-GFP polyclonal antibody (GFP-FL, Santa Cruz Biotechnology), a rabbit anti-PARP-p85-fragment polyclonal antibody (Promega; this antibody does not detect the p116 fragment of intact PARP)] and 1:2000 [a mouse anti-FLAG monoclonal antibody (FLAG-M5, SIGMA), mouse anti-β-actin monoclonal antibody (clone AC-74, SGMA)]. After several washings with TBST, the membrane was incubated with secondary antibody (1:2000, horseradish-peroxidase-labeled anti-rabbit or anti-mouse IgG, Amersham) for 1 h at 20°C, followed by repeat washing in TBST. Reaction products were visualized with the ECL kit (Amersham). Western blots for PARP-p85, which is used as a surrogate for apoptosis, were repeated twice each for samples obtained from three independent experiments. Western blot for β-actin was used as a loading control. The resultant bands for PARP-p85 and β-actin were quantified using the Scion Image processing and analysis program (Scion Corporation) and expressed as IDVs. Each PARP-p85-IDV was normalized to β-actin-IDV as a PARP-p85-IDV/β-actin-IDV (PARP-p85/β-actin) ratio. The ratios obtained from three independent experiments for one distinct group were finally averaged as the mean±SEM using the SPSS statistical software (SPSS Japan) for statistical analyses. A significance level was calculated using the Fisher's post hoc least significant difference test with an assigned confidence interval of 95%.
Immunoprecipitation
Cells cultured in six-well plate were collected 48 h after transfection of GFP-HAP1 and FLAG-ARQ25 or -ARQ65. The aforementioned western blot was performed with ARN-20 or GFP-FL antibodies for 10% of the samples before immunoprecipitation in order to estimate amounts of the exogenously expressed proteins. For the immunoprecipitation, cells from the same sample were disrupted in 3 ml of a lysis buffer (5 m M EDTA, 0.5% Triton-X) for 1 h at 4°C with gentle shaking and centrifuged at 3000 rpm for 20 min at 4°C to remove debris. The cleared lysate was preincubated with protein A/G-agarose beads (Santa Cruz Biotechnology) prewashed with lysis buffer for 1 h at 4°C. The AR and AR-associated proteins were immunoprecipitated with the mixture of ARN-20 antibody (5 µg) and prewashed-protein A/G-agarose beads (30 µl) at 4°C for 10 h. The complex of AR, AR-associated proteins and beads were collected by centrifugation and washed five times in phosphate-buffered saline (PBS) and resuspended in 30 µl of SDS-sample buffer. Each 10 µl aliquot of the immunoprecipitated sample was subjected to SDS/PAGE and then to western blot with ARN-20 or GFP-FL antibodies. The experiments were repeated three times for samples obtained from two independent experiments.
Immunocytochemistry
HEp-2 cells were fixed with 4% paraformaldehyde in PBS for 20 min, washed three times in PBS and permeabilized in 0.5% Triton X in PBS for 5 min at room temperature. They were then blocked with 5% normal goat serum (NGS) in PBS at room temperature for 1 h, incubated with ARN-20 (1:1000) or FLAG-M5 (1:2000) for 1 h at 20°C and washed in PBS three times, followed by incubation with secondary antibody (Alexa Fluor 594-conjugated goat anti-rabbit or -mouse IgG, Molecular Probes; diluted 1:500) for 1 h at 20°C.
Cell count and statistical analysis
Immunostained cells were analyzed by the image-analysis-software program, Meta Morph (Universal Imaging Corp). Subcellular morphology of AR has also previously been reported to vary even in DHT-free or charcoal-stripped medium ( 68–70 ); some have AR both in nucleus and cytoplasm as inclusions, and others show diffused AR pattern extending over nucleus and cytoplasm. Although AR-expressing cells were previously classified into five phases on the basis of AR-expression patterns ( 69 ), we have not successfully done this. Thus, only the HAP1/AR-cotransfected cells and HA-CAF-cells were counted and HA-CAF ratio (HA-CAF cell number/total cotransfected cell number) was used as a surrogate for an index of the association frequency or strength of AR with HAP1. This study focused on comparison between the indexes for ARQ25 and ARQ65 or for low and high levels of DHT. The cotransfected cells were judged by either diffused or aggregated immunoreactions in nuclei or cytoplasm for AR (Alexa594-red) and HAP1 (GFP-green) in fluorescent microscopy, whereas HA-CAF cells were judged by the double-immunostained cytoplasmic aggregations. Their numbers were counted in a blind manner by two members in our study group in six fields (650×450 µm grid at a magnification of 20×) chosen randomly from each well of triplicated samples prepared in a 48-well plate (total 18 fields per sample). The data were analyzed using Mann–Whitney's U test, with P <0.05 indicating statistical significance.
ACKNOWLEDGEMENTS
We cordially thank Professor Keith R. Yamamoto (School of Medicine, University of California, San Francisco, USA) for generously gifting us the plasmids of human ARQ25, ARQ65, ARN127(25) and ARN127(65), and Dr John Pearson for his critical reading of the manuscript. The authors are grateful to Mr Yutaka Suetomi and Dr Motokazu Koga for the technical assistance in early stages of the present study. We would also like to acknowledge the technical expertise of the DNA Core facility of the Center for Gene Research, Yamaguchi University, supported by a Grant-in-Aid from the Ministry of Education, Science, Sports and Culture of Japan. This study was supported by Grant-in-Aid for Scientific Research (C) (No. 17500231) from the Japan Society for the Promotion of Science (JSPS) and Grant-in-Aids for Young Scientists (B) (No. 17700333, 15790624) and for Scientific Research on Priority Areas (No. 17052016) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT).
Conflict of Interest statement . None declared.