Article Text
Abstract
Introduction Previous reports in European populations demonstrated the existence of five data-driven adult-onset diabetes subgroups. Here, we use self-normalizing neural networks (SNNN) to improve reproducibility of these data-driven diabetes subgroups in Mexican cohorts to extend its application to more diverse settings.
Research design and methods We trained SNNN and compared it with k-means clustering to classify diabetes subgroups in a multiethnic and representative population-based National Health and Nutrition Examination Survey (NHANES) datasets with all available measures (training sample: NHANES-III, n=1132; validation sample: NHANES 1999–2006, n=626). SNNN models were then applied to four Mexican cohorts (SIGMA-UIEM, n=1521; Metabolic Syndrome cohort, n=6144; ENSANUT 2016, n=614 and CAIPaDi, n=1608) to characterize diabetes subgroups in Mexicans according to treatment response, risk for chronic complications and risk factors for the incidence of each subgroup.
Results SNNN yielded four reproducible clinical profiles (obesity related, insulin deficient, insulin resistant, age related) in NHANES and Mexican cohorts even without C-peptide measurements. We observed in a population-based survey a high prevalence of the insulin-deficient form (41.25%, 95% CI 41.02% to 41.48%), followed by obesity-related (33.60%, 95% CI 33.40% to 33.79%), age-related (14.72%, 95% CI 14.63% to 14.82%) and severe insulin-resistant groups. A significant association was found between the SLC16A11 diabetes risk variant and the obesity-related subgroup (OR 1.42, 95% CI 1.10 to 1.83, p=0.008). Among incident cases, we observed a greater incidence of mild obesity-related diabetes (n=149, 45.0%). In a diabetes outpatient clinic cohort, we observed increased 1-year risk (HR 1.59, 95% CI 1.01 to 2.51) and 2-year risk (HR 1.94, 95% CI 1.13 to 3.31) for incident retinopathy in the insulin-deficient group and decreased 2-year diabetic retinopathy risk for the obesity-related subgroup (HR 0.49, 95% CI 0.27 to 0.89).
Conclusions Diabetes subgroup phenotypes are reproducible using SNNN; our algorithm is available as web-based tool. Application of these models allowed for better characterization of diabetes subgroups and risk factors in Mexicans that could have clinical applications.
- insulin resistance
- type 2 diabetes mellitus
- ethnic groups
- statistical models
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
Significance of this study
What is already known about this subject?
Previous research by European groups demonstrated that data-driven adult-onset diabetes subgroups have significant clinical and outcome-related implications, with similar patterns and consistency across studies.
Estimation of insulin action using C-peptide-based homeostasis model assessment measures and assessing glycemic control based on HbA1c might limit the access to clustering solutions using unsupervised learning.
What are the new findings?
Reproducibility of diabetes subgroup classification is significantly improved using self-normalizing neural networks.
Application of these models in Mexican cohorts allowed us to characterize differences in diabetes subgroup frequencies, risk factors, chronic complications, clinical trajectories, metabolic and genetic traits.
Diabetes subgroup classification could be useful for treatment selection and, if repeated after interventions, might be useful in identifying groups at higher risk for complications.
How might these results change the focus of research or clinical practice?
Our study shows that diabetes subgroups can be used to understand specific traits of diabetes and improve personalized medicine.
Our approach may lead to wider use of this subgroup classification in more diverse research settings to address heterogeneity of diabetes in different ethnic groups.
Introduction
Recent reports in European populations described a novel classification of adult-onset diabetes mellitus with implications for the prediction of outcomes and disease progression.1 2 Classification of these subgroups uses unsupervised k-means clustering based on six variables: autoantibodies associated with autoimmune diabetes, age at diabetes diagnosis, glycated hemoglobin (HbA1c), homeostasis model assessment (HOMA)2-IR and HOMA2-β estimated using C-peptide and the body-mass index (BMI). Data-driven diabetes subgroups could be useful in admixed populations where heterogeneity of diabetes remains unaddressed because of lack of resources or awareness. Furthermore, most metabolic traits associated with each subgroup including insulin resistance (IR), adipose tissue function and ectopic fat accumulation have ethnic-specific differences which may modify behavior of these subgroups in non-European populations.3 4 Additional efforts to address diabetes heterogeneity using genetic and clinical markers have been explored; nevertheless, the applicability of these approaches remains unclear and its complexity might limit its application in lower resource settings.5 6 Admixed populations, such as Mexico, are highly heterogeneous and the prospect of using these approaches to identify specific traits of diabetes for personalized medicine is appealing.
The k-means clustering algorithm previously used for diabetes subgroup classification is robust. However, its reproducibility depends on initial centroid value seeding, data ordering, extreme outliers and variance of clustering variables.7–9 Low reproducibility might be a concern in settings where C-peptide or HbA1c measurements are limited and clustering is carried out using surrogate insulin or non-insulin-based alternatives to estimate insulin action. To translate the concept of diabetes subgroups to more diverse research settings, we propose a supervised machine learning (ML) approach using artificial self-normalizing neural networks (SNNNs) trained with surrogate metabolic measures to estimate insulin action-related phenomena from population-based studies. SNNN is an ML algorithm which addresses variance by processing the inputs through self-normalizing layers, offering higher precision for classification and regression tasks compared with other ML models.10 We hypothesized that training SNNN to classify diabetes subgroups would improve reproducibility of this approach in independent datasets and would then be useful to characterize diabetes traits in Mexicans. Once we trained the algorithm, we applied it to four Mexican cohorts to understand aspects of diabetes at different stages for the disease including risk factors for diabetes subgroup incidence, nationally representative subgroup prevalence, clinical management and response to treatment during clinical follow-up as well as risk for chronic complications.
Methods
National Health and Nutrition Examination Survey (NHANES) cohorts
NHANES is a population-based survey. It aims to collect information on clinical and health data in a representative multiethnic sample in the USA. We extracted data from four NHANES survey cycles: 1988–1994 (NHANES-III), 1999–2000, 2001–2002 and 2003–2004, including subjects previously diagnosed with type 2 diabetes (T2D) for <5 years, with a HbA1c >6.5% and/or a 2-hour plasma glucose >200 mg/dL following a 75 g oral glucose tolerance test. Subjects were assumed to be anti-Glutamate decarboxylase (GAD65) negative, since measurement of this antibody had only been carried out in a subset of the population. Based on that, we did not consider the severe autoimmune diabetes (SAID) subtype in our estimations.11 The homeostasis model assessment was used to estimate HOMA2-IR and HOMA2-β using fasting plasma glucose (FPG) and C-peptide or fasting insulin for HOMA2-IR’ and HOMA2-β’.
Artificial SNNNs
We fitted four SNNN models to develop a classification algorithm for diabetes subgroups:
Model 1: HOMA2-IR, HOMA2-β, BMI, HbA1c, years since diagnosis.
Model 2: HOMA2-IR’, HOMA2-β’, BMI, HbA1c, years since diagnosis.
Model 3: HOMA2-IR’, HOMA2-β’, BMI, FPG and years since diagnosis.12
Model 4: Replacing HOMA for METS-IR (a non-insulin-based model for IR, metabolic score for IR), METS-VF (a visceral fat estimator, metabolic score for visceral fat),13 14 HbA1c, BMI and age at diabetes onset.
Performance and fine-tuning of SNNN models were assessed with cross-validation (k=10) and in a validation sample (NHANES 1999–2004).
Analytical approach for SNNN algorithm testing cohorts
Once we trained these models and verified the reproducibility of diabetes subgroups, we aimed to investigate specific traits of diabetes regarding risk factors for subgroup incidence, subgroup prevalence, clinical trajectories and risk for chronic complications in four Mexican cohorts. Complete description of these cohorts is included in online supplementary material.
Supplemental material
Risk factors for diabetes subgroup incidence
To investigate these factors, we used the Metabolic Syndrome (MS) cohort (n=6144), an open-population study developed to evaluate the risk of incident T2D and cardiovascular disease in urban populations living in nine different Mexican cities.15 Subjects were assessed to obtain medical history, physical activity habits and anthropometric/biochemical analyses. These same evaluations were carried out after a minimum of 2 years of follow-up. We search for risk factors associated with the incidence of each diabetes subgroup; for this purpose, we used competing risk analyses using the survival R package. Diabetes subgroups in the MS cohort were classified using SNNN model 3 due to the unavailability of HbA1c and fasting C-peptide.
Diabetes subgroup national prevalence estimates
To estimate diabetes subgroup prevalence, we used data collected from ENSANUT 2016 Medio Camino (n=4023), a nationally representative survey to evaluate nutrition and health trends in Mexicans in whom blood samples were collected for subgroup classification. Subjects with previous diagnosis of diabetes, HbA1c ≥6.5% or FPG ≥126 mg/dL were included in this analysis. Prevalence and 95% CIs were constructed considering multistage-stratified and clustered sampling using the survey R package.16 Diabetes subgroups in ENSANUT 2016 were classified using SNNN model 2 using insulin-based surrogates for HOMA2-IR and HOMA2-β.
Chronic complication profiles and diabetes subgroups
To evaluate these profiles, we analyzed the SIGMA-UIEM cohort (n=1521), an open-population study designed to characterize carriers and non-carriers of SLC16A11 variants associated with increased risk for T2D in Mexicans. In a subset of subjects, we assessed the presence of diabetic kidney disease (DKD) using the albumin to creatinine ratio, diabetic neuropathy (DN) using the Michigan questionnaire (n=1123) and diabetic retinopathy (DR) using a standardized ophthalmological examination (DR, n=353). To assess non-alcoholic fatty liver disease (NAFLD), we used the fatty liver index (FLI).17 Risk for chronic complications and associations of diabetes subgroups with the SLC16A11 variant were assessed using propensity score-matched analyses, controlling for years from diabetes diagnosis, age and sex using logistic mixed-effects models. A subsample of study participants (n=67) underwent deep phenotyping (online supplementary material).18 19 Insulin sensitivity was assessed using raw, weight and insulin-adjusted M-values from euglycemic hyperinsulinemic clamps (EHCs). To evaluate acute insulin response to glucose (AIRg), a frequently sampled intravenous glucose tolerance test was performed. Subcutaneous and visceral adipose tissue areas (SFA, VFA) were quantified using MRI, and intrapancreatic and intrahepatic triglyceride contents were determined using MRI spectroscopy. Diabetes subgroups in the SIGMA-UIEM cohort were classified using SNNN model 2.
Clinical follow-up of diabetes subgroups
Clinical assessments for each diabetes subgroup were evaluated using data from the CAIPaDi cohort (n=1608), an open-population multidisciplinary diabetes management program (online supplementary material).20 For this evaluation we included subjects who completed follow-up at 3 months, 1 and 2 years. Diabetes subgroup classification was conducted at baseline and at 3 months, 1 and 2 years after the original intervention to assess diabetes-subgroup transitions across time. We evaluated treatment response using Cox proportional risk regression models and assessed individual mediation groups according to HbA1c goal attainment after follow-up for each diabetes subgroup.
Statistical analysis
Descriptive statistics are reported as mean±SD or as median±IQR, where appropriate. Missing data were imputed using multivariable imputation with chained equations when data were missing at random using the mice R package. Specific traits of diabetes subgroups in all evaluated cohorts were compared using one-way analysis of variance or Kruskal-Wallis test with post hoc Tukey or Dunn test. Paired measures in the MS cohorts were compared using paired t-test or Wilcoxon test, where appropriate. Statistical significance was established at a two-tailed p-value <0.05; all statistical analyses were carried out using R 3.6.1.
Diabetes subgroup clustering
For diabetes subgroup classification in NHANES, we standardized HOMA2-IR, HOMA2-β, HbA1c, BMI and age at diagnosis into z-scores and performed k-means clustering using the fpc R package (k=4, 100 runs). As previously described, four subgroups were identified1 2 11 12: severe insulin-deficient diabetes (SIDD), severe insulin-resistant diabetes (SIRD), mild obesity-related diabetes (MOD) and mild age-related diabetes (MARD). We hypothesized that using surrogate variables instead of original clustering inputs would impact classification accuracy; to test this hypothesis, we performed the k-means clustering algorithm substituting C-peptide variables and HbA1c using variable combinations as described for models 2–4 and compared these results to classification using fully trained SNNN models 2–4. To evaluate reproducibility of SNNN compared with k-means clustering using surrogate variables, we used confusion matrices and areas under receiver operating characteristic curves.
Diabetes subgroup incidence and prediction
To investigate risk factors for diabetes subgroup incidence, we matched cases of diabetes with controls using propensity score matching for age, sex and BMI with the MatchIt R package. Risk factors were modeled using Fin & Gray semiparametric competitive risk regression to account for competing risks between subgroups, adjusted for age, sex, waist circumference, smoking, family history of diabetes and physical activity to account for residual confounding.
Diabetes complications, genetic associations, clinical trajectories and cluster transitions
To investigate the association of diabetes subgroups with chronic complications in the UIEM-SIGMA and CAIPaDi cohorts, we used fixed-effects logistic regression adjusted for sex and years since T2D diagnosis. Genetic associations for the SLC16A11 risk variant were assessed using mixed-effects logistic regression models in propensity score matched individuals for sex, years from T2D diagnosis and HbA1c. For prospective evaluations in CAIPaDi, we modeled risk using Cox regressions excluding prevalent DKD and DR cases. To assess subject transitions in diabetes subgroups across time, we used Sankey plots and confusion matrices; the validity of diabetes subgroups at baseline and its transitions or stability at 3 months after the intervention for predicting metabolic trajectories and risk of chronic complications were also assessed. Finally, to explore the effect of medications in reaching glycemic targets (HbA1c <7.0%) after 3 months and 1 year, we used Cox proportional risk regression analyses, introducing treatment by diabetes subgroup and treatment by subgroup transition interactions to investigate specific effects of medications by cluster.
Results
Diabetes clusters in NHANES
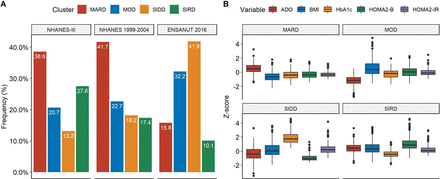
For diabetes subgroup classification, we merged the NHANES-III (n=20 050) and NHANES 1999–2004 datasets (n=41 470). Of the 1865 subjects with <5 years of diabetes diagnosis, 63 had incomplete data and 44 additional subjects who had data >5 SD from the mean were eliminated from the analysis. In those remaining (n=1758) we performed a k-means clustering algorithm using C-peptide derived measures, HbA1c, years from diabetes diagnosis and BMI as described in previous diabetes clustering studies. These groups showed similar distributions in both NHANES III and 1999–2004 NHANES; clinical variables followed expected patterns for each subgroup, including surrogate measures (figure 1; online supplementary figure 1). SNNN models were trained for 50 epochs using NHANES-III (n=1132) and validated in NHANES 1999–2004 (n=626).
(A) Diabetes subgroup distribution in NHANES III used for model training, NHANES 1999–2004 used for model validation and ENSANUT 2016 used for model testing, demonstrating relevant differences in diabetes distribution. (B) Distribution of type 2 diabetes clusters according to ADO, HOMA2-β, HOMA2-IR, BMI, HbA1c and fasting plasma glucose in the combined NHANES cohorts. ADO, age at diabetes onset; BMI, body mass index; HbA1c, glycated hemoglobin; HOMA, homeostasis model assessment; IR, insulin resistance; MARD, mild age-related diabetes; MOD, mild obesity-related diabetes; NHANES, National Health and Nutrition Examination Survey; SIDD, severe insulin-deficient diabetes; SIRD, severe insulin-resistant diabetes.
Performance of SNNN models and comparison with k-means clustering
SNNN model 1 showed excellent classification performance. With both SNNN models 2 and 4, the classification performance ordered from better to worse was SIDD, MOD, MARD and SIRD, with the greatest misclassification occurring between MARD and SIRD, compared with the original clustering results (table 1). With SNNN model 3, the order of better to worsening performance was MOD, followed by SIDD, MARD and SIRD. Diagnostic performance measures for all SNNN models were significantly improved compared with k-means unsupervised clustering using variable combinations from models 2–4 (online supplementary tables 2 and 3). To facilitate the use of these models, we deployed them into an external web-based interactive tool built using the shiny R package, which is accessible for researchers and clinicians at: https://uiem.shinyapps.io/diabetes_clusters_app/.
Performance of the four trained SNNN models contrasting classification metrics k-fold cross-validation (k=10) of the SNNN algorithm and the performance in the testing (NHANES 1999–2004) and training datasets (NHANES-III)
Diabetes subgroup prevalence in the population-based nationwide survey
Clinical variables followed the expected pattern for each subgroup in all cohorts (online supplementary figures 2–7). In ENSANUT 2016, we observed a high prevalence of the insulin-deficient form (41.25%, 95% CI 41.02% to 41.48%), followed by obesity-related diabetes (33.60%, 95% CI 33.40% to 33.79%), age-related diabetes (14.72%, 95% CI 14.63% to 14.82%) and severe insulin-resistant groups (10.43%, 95% CI 10.33% to 10.53%; figure 1). Overall, insulin-deficient cases were more likely to have >5 years since diabetes diagnosis. Women had higher rates of age-related diabetes, and there was a higher-than-expected rate of the severe insulin-resistant forms in Mexico City. No other subgroup had significant differences in its distribution by either gender, urban/rural setting or geographical area (table 2).
Population-based prevalence and 95% CI estimates of diabetes subgroups in Mexican population based on ENSANUT 2016 data after application of the SNNN algorithm (n=614, representing 8 487 590 Mexicans), comparing different subgroups related to years since diagnosis, setting, sex and geographical region
Diabetes subgroup deep phenotyping in the SIGMA-UIEM cohort
The detailed characterization done in the SIGMA-UIEM cohort provided confirmatory evidence of the metabolic derangements expected in each diabetes subgroup. EHC-derived raw and insulin-adjusted M-values were lower in SIRD subjects confirming the insulin-resistant phenotype. AIRg was lower for SIDD implying reduced β-cell response; conversely, MOD/SIRD had enhanced AIRg indicating response to systemic IR. Regarding fat distribution, SFA and the SFA/VFA ratio were higher in MOD confirming predominance of subcutaneous adiposity. Subjects with MOD also had higher fat mass by Dual X-ray absorciometry (DXA) and SIRD had lower total lean mass and lower bone mineral content compared with MOD/MARD. Intrahepatic fat was surprisingly lower in SIRD compared with MOD/SIDD (online supplementary table 4).
We searched for associations between diabetes subgroups and the risk variant for SLC16A11 using mixed-effects logistic regression models with propensity score matching for sex, HbA1c and years of T2D exposure. We observed a significant association between this variant and the MOD subgroup in the carrier status analyses (OR 1.42, 95% CI 1.10 to 1.83, p=0.008) and even comparing heterozygous (OR 1.41, 95% CI 1.07 to 1.85, p=0.013) and homozygous status (OR 1.44, 95% CI 1.01 to 2.076, p=0.048).
Risk for incident diabetes subgroups in the MS cohort
In the MS cohort, after a median of 2.3 years of follow-up we observed 331 cases of incident T2D; among them, we observed a greater incidence of MOD (n=149, 45.0%), followed by SIRD (n=118, 35.6%), MARD (n=45, 13.6%) and SIDD (n=19, 5.7%). Using competing risks regression, we identified that adults >60 years with inappropriately low HOMA2-β, normal BMI, who used statins and were physically inactive had higher risk for MARD. Subjects <40 years old, with elevated HOMA2-IR/HOMA2-β and metabolic syndrome (MS) by International Diabetes Federation (IDF) criteria had higher risk for MOD. Subjects at-risk of SIRD had elevated HOMA2-IR/HOMA2-β, were older compared with MOD but younger than MARD and had MS by Adult Treatment Panel III (ATP-III) criteria. Finally, subjects at-risk of SIDD already had inappropriately lower HOMA2-β despite higher HOMA2-IR values (online supplementary table 5; table 3).
Fine & Gray semiproportional hazard regression for diabetes subgroup using competing risk between subgroups to identify factors associated to diabetes subgroup incidence in Mexican population compared with age, sex and BMI propensity score matched controls (n=991), adjusted for family history of diabetes, physical activity, waist circumference, smoking, age and stratified by sex
Association of diabetes subgroups with chronic diabetes complications
In the SIGMA-UIEM cohort, we identified a lower prevalence of chronic complications for obesity-related diabetes (particularly retinopathy and nephropathy; online supplementary table 6). Subjects with MARD had decreased risk of DN and NAFLD only. Subjects with SIDD had increased risk of DKD, NAFLD and DR. SIRD was associated with higher risk for NAFLD and estimated glomerular filtration rate (eGFR) <60 mL/min (online supplementary table 7).
In the CAIPaDi cohort, lower risk for DR rates were observed for MOD (OR 0.59, 95% CI 0.44 to 0.79) and SIRD (OR 0.43, 95% CI 0.18 to 0.85) but the risk was greater in SIDD at baseline (OR 1.90, 95% CI 1.47 to 2.47). DKD rates at baseline were lower for MOD (OR 0.40, 95% CI 0.16 to 0.86) and MARD (OR 0.62, 95% CI 0.40 to 0.92), but higher for SIDD (OR 4.78, 95% CI 2.29 to 11.24). When excluding prevalent cases, we observed increased 1-year (HR 1.59, 95% CI 1.01 to 2.51) and 2-year risk (HR 1.94, 95% CI 1.13 to 3.31) of incident DR in SIDD and decreased 2-year DR risk for MOD (HR 0.49, 95% CI 0.27 to 0.89) without differences for incident DKD.
HbA1c targets and treatment response according to diabetes subgroup
After 3 months, we observed lower rates of glycemic control achievement (HbA1c <7%) in SIDD (61.6% vs 90.2% in MARD, 92.1% in MOD and 98.0% in SIRD, p<0.001) compared with other subgroups. HbA1c targets remained lower at 1 and 2 years for SIDD (46.3%, 43.3%), followed by MOD (73.1%, 64.5%), MARD (80.0%, 80.9%) and SIRD (90.6%, 87.1%). Overall, subjects with SIDD (HR 0.45, 95% CI 0.32 to 0.83) and MOD (HR 0.68, 95% CI 0.46 to 0.83) were less likely to achieve glycemic control at 2 years compared with MARD.
Association of subgroup transitions with clinical trajectories and treatment response
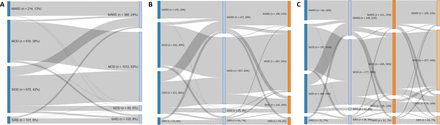
In the CAIPaDI cohort, only 10.7% of SIDD subjects remained in this subgroup and most were reclassified to either MOD (65.8%) or MARD (19.5%) at the 3-month time point, whereas other groups remained relatively stable over time (MARD 97.7%, MOD 91.6% and SIRD 74.2%; figure 2). We re-estimated the risk of chronic complications considering subgroup classification at 3 months and observed that subjects who were SIDD at baseline and remained so had higher 2-year risk of RD (HR 5.80, 95% CI 2.12 to 15.88) and higher 1 year risk of DKD (HR 3.56, 95% CI 1.18 to 10.75) compared with those who transitioned. Clinical trajectories of markers including HbA1c, body fat, FLI and METS-VF also show differential responses for subgroups classified at baseline and at 3 months at different time points, particularly for SIDD and MOD (online supplementary figures 8 and 9; online supplementary table 8).
Sankey plot of transitions of diabetes subtypes after 3 months (A, n=1680), 1 year (B, n=852) and 2 years (C, n=476) of an intensive multidisciplinary intervention with variables collected at baseline and after 3 months, 1 and 2 years of follow-up. MARD, mild age-related diabetes; MOD, mild obesity-related diabetes; SIDD, severe insulin-deficient diabetes; SIRD, severe insulin-resistant diabetes.
Discussion
Clustering of data-driven diabetes subgroups is heavily influenced by variable selection. Using metabolic surrogates yielded low reproducibility of diabetes subgroups, a discrepancy which was corrected for using SNNN models. Our study confirms that SNNN models trained using population-based data can better reproduce diabetes subgroup classification using surrogate measures. Application of these models allowed for the characterization of diabetes subgroups in Mexicans using a unique combination of cohorts, which comprises a wide pathophysiological spectrum ranging prior to diabetes onset, early diagnosis and clinical trajectories, and assessing risk of chronic complications in a heterogeneous population with elevated genetic risk for diabetes.19 Ours is the first attempt to generalize diabetes subgroup classification using surrogate measures by using supervised ML algorithms. Widespread use of ML to improve research in metabolism has led to significant improvements in risk prediction.21 The use of unsupervised clustering is particularly useful in situations where C-peptide and HbA1c measurements are available for subgroup classification. By developing SNNN algorithms trained on clustered data from ethnically diverse cohorts such as NHANES, one is able to minimize the effect of surrogate measure variability in diabetes subgroup classification that unsupervised clustering would otherwise produce, resulting in profiles that are more reproducible in independent cohorts. Our approach could promote application of these subgroups in populations with a variety of risk profiles, in whom large-scale studies with C-peptide or even insulin measurements are unavailable, improving reproducibility at lower costs.
Mexican population is admixed with predominant Amerindian ancestry and a higher risk of T2D compared with European populations.22 The elevated prevalence of T2D in Mexicans is the result of genetic predisposition and an increased prevalence of obesity and MS due to unhealthy lifestyles.23 Unsurprisingly, prevalence of diabetes subgroups in Mexican population did not follow reported patterns from European, US and Chinese cohorts.1 2 11 The larger prevalence of insulin-deficient cases could be attributable to poor metabolic control resulting from health-related disparities and high rates of long-standing undiagnosed diabetes in Mexicans, which was reinforced with our finding of increased SIDD prevalence in subjects with >5 years of disease and by considering that incidence for SIDD was low, despite higher risk profiles in the MS Cohort. Impaired β-cell function could result from glucotoxicity in uncontrolled diabetes, which could be reversed with prompt and adequate treatment.24 The large increase in MOD/MARD and the drastic reduction in SIDD prevalence after a 3-month multidisciplinary intervention to improve glycemic control showed that most SIDD cases were transient.
In contrast with prevalence data, we also reported a large incidence of obesity-related and insulin-resistant cases, possibly influenced by the more adverse risk profile of study participants. A high prevalence of MS, hypoalphalipoproteinemia and abdominal obesity as well as earlier diabetes onset have previously been reported in Mexicans.21 23 Mexicans are more susceptible to ectopic and visceral fat accumulation, resulting in an increased cardiometabolic risk profile, which increases the risk of chronic complications,23 including DKD and NAFLD, both of which are primarily associated with SIDD/SIRD and MOD, respectively.2 11 Our data show that diabetes subgroup classification could lead to better treatment selection and risk profiling for chronic complications and support the idea that diabetes phenotypes are dynamic and should be reassessed periodically to understand clinical trajectories and reassess the risk of personalized medicine.
A potential limitation of our approach is the exclusion of the SAID subgroup. Adult-onset autoimmune diabetes usually presents with acute diabetes-related complications and poor metabolic control, which increases clinical suspicion and prompts autoantibody testing, despite measures of autoantibodies varying over time. Since ML methods rely on non-readily observed patterns between variables, the use of a variable which is definitive to establish a subgroup does not benefit from this approach. Instead, future efforts to characterize and improve SAID prediction should focus on predicting who might require antibody testing and its heterogeneity might be addressed from independent cluster analysis, as has been carried out for type 1 diabetes.25 26 Finally, previous reports have suggested that autoimmune diabetes has a lower prevalence and incidence in the Mexican population compared with other populations which reduces the likelihood of undiagnosed SAID cases in our population.27 The inclusion of diverse cohorts is a robust approach; however, given that these studies were not specifically designed to investigate factors related to diabetes subgroups, the results require additional replication in independent datasets. Given the flexibility of our approach, this can be performed using a wide variety of available datasets.
Conclusion
The use of SNNN improves reproducibility of diabetes subgroups when using surrogate measures to estimate insulin action compared with unsupervised clustering. Our approach is particularly useful in populations in limited resource settings, in large-scale epidemiological studies lacking the original clustering variables or in primary-care settings in which most of these measures are unavailable. Traits diabetes subgroups identified by the SNNN algorithm are consistent with the distinctiveness of diabetes in Mexicans, and the novel risk profiles and differential treatment responses might significantly impact clinical practice. Further applications of this approach could further characterize ethnic-specific traits associated with diabetes in ours and other populations. Diabetes subgroups are a promising approach to permit the application of personalized medicine in diabetes. By improving its reproducibility, we hope to better understand the clinical relevance of this classification in more diverse research settings.
Acknowledgments
All authors approved the submitted version. All the authors thank the staff of the Endocrinology and Metabolism Department for all their support, particularly to Luz Elizabeth Guillen-Pineda, Maria Del Carmen Moreno-Villatoro and Adriana Cruz-Lopez. We are thankful to the study volunteers for all their work and support throughout the realization of the study.
References
Footnotes
Collaborators The Metabolic Syndrome Study Group: Olimpia Arellano-Campos, Donaji V Gómez-Velasco, Omar Yaxmehen Bello-Chavolla, César Lam-Chung, Ivette Cruz-Bautista, Marco A Melgarejo-Hernandez, Paloma Almeda-Valdés, Alexandro J Martagón, Liliana Muñoz-Hernandez, Luz E Guillén, José de Jesús Garduño-García, Ulices Alvirde, Yukiko Ono-Yoshikawa, Ricardo Choza-Romero, Leobardo Sauque-Reyna, Ma Eugenia Garay-Sevilla, Juan M Malacara-Hernandez, María Teresa Tusié-Luna, Luis Miguel Gutierrez-Robledo, Francisco J Gómez-Pérez, Rosalba Rojas, Carlos A Aguilar-Salinas. Group of Study CAIPaDi: Sergio Hernández-Jiménez, Cristina García-Ulloa, Eder Patiño-Rivera, Denise Arcila-Martínez, Rodrigo Arizmendi-Rodríguez, Oswaldo Briseño-González, Humberto Del Valle-Ramírez, Arturo Flores-García, Fernanda Garnica-Carrillo, Eduardo González-Flores, Mariana Granados-Arcos, Héctor Infanzón-Talango, Victoria Landa-Anell, Claudia Lechuga-Fonseca, Arely López-Reyes, Marco Melgarejo-Hernández, Angélica, Palacios-Vargas, Liliana Pérez-Peralta, Alberto Ramírez-García, David Rivera de la Parra, Sofía Ríos-Villavicencio, Francis Rojas-Torres, Marcela Ruiz-Cervantes, Sandra Sainos-Muñoz, Alejandra Sierra-Esquivel, Erendi Tinoco-Ventura, Luz Elena Urbina-Arronte, María Luisa Velasco-Pérez, Héctor Velázquez-Jurado, Andrea Villegas-Narváez, Verónica Zurita-Cortés, Aída Jiménez-Corona, Enrique Graue-Hernández, Carlos Aguilar-Salinas, Francisco J Gómez-Pérez, David Kershenobich-Stalnikowitz.
Contributors Research idea and study design: OYB-C, JPB-L, AV-V, NEA-V, RM, PA-V, CAA-S; data acquisition: PA-V, RM, AV-V, IC-B, RR, SH-J, ACG-U,CAA-S; data analysis/interpretation: OYB-C, JPB-L, CAA-S; statistical analysis and machine learning: OYB-C; manuscript drafting: OYB-C, JPB-L, AV-V, NEA-V, SH-J, ACG-U, RR, RM, PA-V, IC-B, CAA-S; supervision or mentorship: CAA-S. Each author contributed important intellectual content during manuscript drafting or revision and accepts accountability for the overall work by ensuring that questions pertaining to the accuracy or integrity of any portion of the work are appropriately investigated and resolved.
Funding The SIGMA-UIEM cohorts were conducted as part of the Slim Initiative for Genomic Medicine, a project funded by the Carlos Slim Health Institute in Mexico and the Consejo Nacional de Ciencia y Tecnologia. Grant Infraestructura 255 096. The Metabolic Syndrome cohort was supported by a grant from the “Consejo Nacional de Ciencia y Tecnología (CONACyT)” (S0008-2009-1-115250) and research grant by Sanofi. The CAIPaDi program has received grants from Astra Zeneca, Fundación Conde de Valenciana, Novartis, Consejo Nacional de Ciencia y Tecnología (214718), Nutrición Médica y Tecnología, NovoNordisk, Boehringer Ingelheim, Dirección General de Calidad y Educación en Salud, Eli Lilly, Merck Serono, MSD, Silanes, Chinoin and Carlos Slim Health Institute.
Disclaimer The funding bodies had no roles in the design of the study and collection, analysis, interpretation of data and in writing the manuscript. The sponsors had no role in the conception, development, analyzing, writing or editing of this document.
Competing interests JPB-L, AV-V and NEA-V are enrolled at the PECEM program of the Faculty of Medicine at UNAM. JPB-L and AV-V are supported by CONACyT.
Patient consent for publication Not required.
Provenance and peer review Not commissioned; externally peer reviewed.
Data availability statement Data are available upon reasonable request to the corresponding author.