Article Text
Abstract
Background Type 2 diabetes mellitus (T2DM) is a multifactorial disorder that leads to alterations in gene regulation. Long non-coding RNAs (lncRNAs) have become a major research topic as they are involved in metabolic disorders.
Methods This study included a total of 400 study subjects; 200 were subjects with T2DM and 200 were healthy subjects. Extracted RNA was used to synthesize cDNA by quantitative real time. Serum analysis was carried out to determine differences in biochemical parameters. Recorded data were used to evaluate associations with expression of lncRNAs NF-kappaB interacting lncRNA (NKILA), nuclear enriched abundant transcript 1 (NEAT1), metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), and myocardial infarction-associated transcript (MIAT) in T2DM cases.
Results Compared with healthy controls, patients with T2DM showed an overall increase in expression of lncRNAs NKILA, NEAT, MALAT1, and MIAT by 3.94-fold, 5.28-fold, 4.46-fold, and 6.35-fold, respectively. Among patients with T2DM, higher expression of lncRNA NKILA was associated with hypertension (p=0.001), smoking (p<0.0001), and alcoholism (p<0.0001). Altered NEAT1 expression was significantly associated with weight loss (p=0.04), fatigue (p=0.01), slow wound healing (p=0.002), blurred vision (p=0.008), loss of appetite (p=0.007), smoking (p<0.0001), and alcoholism (p<0.0001). Higher expression of lncRNA MALAT1 was significantly linked with weight loss (p=0.003), blurred vision (p=0.01), smoking (p<0.0001), and alcoholism (p<0.0001). Expression of lncRNA MIAT was associated with only blurred vision (p<0.0001), smoking (p<0.0001), and alcoholism (p<0.0001). Positive correlations of lncRNA NKILA with lncRNAs NEAT1 (r=0.42, p<0.0001), MALAT (r=0.36, p<0.0001) and MIAT (r=0.42, p<0.0001) were observed among patients with T2DM. Significant positive correlations of lncRNA NEAT with lncRNAs MALAT and MIAT were observed among patients with T2DM. A positive correlation between lncRNAs MALAT and MIAT was also observed among patients with T2DM.
Conclusion Increased circulating NKILA, NEAT1, MALAT, and MIAT expression in patients with T2DM, which is linked with poor patient outcomes and significantly linked with alcoholism and smoking, may influence the degree and severity of disease among patients with T2DM. These lncRNAs may contribute to the progression of T2DM disease or other related diabetes-related complications.
- diabetes mellitus
- type 2
- gene expression
- RNA
- blood pressure
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
Significance of this study
What is already known about this subject?
Long non-coding RNAs (lncRNAs) have increasingly come into the spotlight due to their increasing functional importance in gene regulation and involvement in health and disease conditions.
T2DM impairs critical functions such as viability, protective mechanisms against oxidative stress, as well as secretory capacity, suggesting that it leads to impaired glucose homeostasis and insulin sensitivity through the alteration in non-coding RNA (ncRNA) expression as well as apoptosis; mitochondria deterioration also participated in the development of T2DM.
So far as per our literature search, none has explored the circulating NF-kappaB interacting lncRNA (NKILA), nuclear enriched abundant transcript 1 (NEAT1), metastasis-associated lung adenocarcinoma transcript 1 (MALAT), and myocardial infarction-associated transcript (MIAT) in the Indian population.
Significance of this study
What are the new findings?
Increased circulating NKILA, NEAT1, MALAT, and MIAT expression was observed in patients with T2DM.
Increased expression was significantly linked with alcoholism, and smoking may influence the degree and severity of disease among patients with T2DM.
Overexpression of the lncRNAs NKILA, NEAT1, MALAT, and MIAT showed a positive correlation, and together, these lncRNAs may contribute to the progression of T2DM or contribute to other diabetes-related severe complications, which could be life-threatening.
A better understanding of underlying the functions and mechanisms of lncRNAs will assist us in recognizing the pathophysiology of diabetes and its related difficulties, and thereby, these could be used to adapt treatment strategies and to develop novel therapeutic approaches shortly.
How might these results change the focus of research or clinical practice?
The present study provides the benchmark to explore the circulating ncRNA because they are released from the specific organ due to cellular apoptosis in a disease condition.
During the progression of diabetes, loss of the pancreatic beta-cell by apoptosis as well as in other related complications during the time cells release the nucleic acid in circulation and that can be used as a prognostic marker in patients’ management.
Introduction
Diabetes mellitus (DM) is a clutch of chronic metabolic disorders characterized by hyperglycemia and impaired insulin oozing or function.1 The incidence of DM is aggregating worldwide, and it was predicted that global dominance will increase from 415 million cases in 2015 to 642 million cases by 2040.2 Type 2 diabetes mellitus (T2DM) is a complex, multifactorial condition that arises from combined interactions between genetic/epigenetic and environmental factors.3 Long non-coding RNAs (lncRNAs), longer than 200 nucleotides, the leading class of non-coding RNAs (ncRNAs).4 Recent research has conveyed that lncRNA expression profiles are misregulated in many human diseases.5 Also, genome-wide association study showed that several genetic alterations within lncRNAs are associated with disease susceptibility.6 Alterations in genes have been shown to affect the expression levels of lncRNAs.7 LncRNAs have increasingly come into the spotlight due to increasing their functional importance in both health and disease conditions.8 LncRNAs are the most important protein-coding genes that tightly control transcriptional, and their expression pattern often connects with diseases and in tissue differentiation.9
Apart from protein-coding genes that contributed to disease development in humans, research has emphasized that lncRNAs actively involved in pathogenesis and disease progression could be the target for therapeutics development.10 Research literature and evidence revealed that lncRNAs involved in glucose homeostasis contribute to diabetes disease progression and related disorder and have shown that Asians are highly insulin resistant11 and more probable to develop T2DM and linked vascular problems than individuals of other ethnicities.12
Post-transcriptional control of mRNA processing is controlled by the complementary sequence’s recognition by lncRNAs13 and promotes mRNA degradation during14 or adjust mRNA translation and splicing.15 lncRNAs diffusely participate in post-transcriptional mRNA processing by interacting and sequestering specific miRNAs.16 lncRNAs have been presented to be involved in many cellular processes and development,17 differentiation,18 and metabolism.19 A wide variety of functions are carried out by lncRNAs and their definite expression patterns, and their dysregulation can promote the appearance of numerous human diseases.
T2DM impairs critical functions, such as viability, protective mechanisms against oxidative stress, as well as secretory capacity, suggesting that this leads to an impaired glucose homeostasis and insulin sensitivity through the downregulation of ncRNAs20 as well as apoptosis, and mitochondria deterioration also participated in development of T2DM.21
Currently, there are no specific biomarkers for diabetes-related diseases or treatment regimens to halt or prevent disease progression. Hence, the need to explore specific biomarkers to diagnose and detect disease development at an early stage is urgent. Therefore, the present study aimed to evaluate lncRNA expression in patients with T2DM.
Materials and methods
Sample collection
This study included a total of 400 study subjects; 200 study subjects were patients with newly diagnosed untreated T2DM, and 200 were healthy subjects. Three millilitres of blood was withdrawn from all the study subjects and collected in plain vials. Fasting as well as postprandial samples were collected in fluoride vials from all 200 subjects with T2DM, and 1 mL of blood was collected in EDTA vials to measure Hba1c. Fasting blood glucose (glucose level≥126 mg/dL) and postprandial glucose (2-hour blood glucose≥200 mg/dL) were monitored and used as criteria for the diagnosis of T2DM. Data were recorded from patients with T2DM with hypertension (systolic blood pressure (BP)≥140 or diastolic BP≥90 mm Hg), increased urination (polyurea by physical examination and urine test of 8, 12 and 24 hours), weight loss (>4 kg of weight loss in the last 3–6 months), fatigue, slow wound healing, blurred vision (fundus/fluorescein angiography), loss of appetite, smoking, and alcoholism for analysis.
Samples collected in plain vials were centrifuged at 1500 rpm to collect the serum and stored at −80°C for further processing.
Total RNA extraction
Stored serum samples from study participants were thawed, and total circulating RNA extraction was performed using TRIzol reagent (Invitrogen, Thermo Scientific, Massachusetts, USA). All samples were then stored at −70°C in 2 mL nuclease-free Eppendorf tubes. The concentration and quality of circulating RNA were determined by measuring the A260/280 ratio using a Nanodrop spectrophotometer.
cDNA synthesis and quantitative real-time PCR (qRT-PCR) to assess lncRNA expression
One hundred nanograms of total circulating RNA extracted from all the study participants was used to synthesize cDNA using a kit (Verso, Thermo Scientific, USA) following the provided protocol. Expression of the circulating lncRNAs NF-kappaB interacting lncRNA (NKILA), nuclear enriched abundant transcript 1 (NEAT1), metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), and myocardial infarction-associated transcript (MIAT) was determined by qRT-PCR using SYBR Green dye with specific primer sequences (online supplemental table 1). The following program was used for qRT-PCR to amplify the lncRNAs NKILA, NEAT1, MALAT1, and MIAT and β-actin in a 20 μL reaction volume: 40 cycles of initial denaturation at 94°C for 40 s; annealing for 40 s at 64°C (NKILA), 64°C (NEAT1), 60°C (MALAT1), 60°C (MIAT) or 64°C (GAPDH); and extension at 72°C for 40 s. A final additional step at 72°C for 5 min to end the reaction and melting curve examination between 35°C and 90°C were performed to confirm target amplification were performed. A control sample without cDNA was included in each experiment, and every reaction was performed in duplicate. Relative quantification by the 2−(ΔΔCT) method was used to compute expression levels of the lncRNAs NKILA, NEAT1, MALAT1, and MIAT.
Supplemental material
Statistical analysis
All the statistical was analyses were performed using GraphPad Prism software V.6.05. The Mann-Whitney U test was performed to calculate the significance of differences among the groups. Analyses of the qRT-PCR results were performed with the relative cycle threshold (Ct) method. Expression levels of the lncRNAs NKILA, NEAT1, MALAT1, and MIAT were calculated by the relative quantification method using 2–(ΔΔCt). Genes whose expression was increased or decreased by more than onefold were considered to be upregulated or downregulated, respectively. All values were normalized relative to the control values, which were set at 1. A p value of less than 0.05 indicated significance.
Results
Demographic and clinical characteristics of patients with T2DM and healthy controls
In brief, the present study included 200 patients with T2DM and 200 healthy control subjects (online supplemental table 2). Among the patients with T2DM, 68% of patients were men and 32% were women, while among the healthy controls, 70% were men and 30% were women; more details are provided in online supplemental table 2.
Expression of the lncRNA NKILA in patients with T2DM
Patients with T2DM exhibited an overall 3.94-fold higher expression of the lncRNA NKILA compared with healthy subjects (table 1). Among the patients with T2DM, significant differences in expression of the lncRNA NKILA were observed among hypertensive and non-hypertensive patients with T2DM (p=0.001). Compared with non-smoking patients with T2DM, smoking patients with T2DM showed a 5.28-fold expression of the lncRNA NKILA, while non-smoking patients with T2DM showed a 2.81-fold change in expression of the lncRNA NKILA, and these values were significantly different (p<0.0001). Compared with healthy subjects, patients with T2DM with alcoholism showed a 5.87-fold change in expression of the lncRNA, while patients with T2DM without alcoholism showed a 1.94-fold change in expression of the lncRNA NKILA; these values were found to be significantly different (p<0.0001).
Association of expression of the circulating lncRNA NKILA with different parameters among patients with T2DM
Expression of the lncRNA NEAT1 in patients with T2DM
Overall, 5.28-fold higher expression of lncRNA NEAT1 was observed in patients with T2DM compared with healthy controls (table 2). Among the patients with T2DM, significant differences in lncRNA NEAT1 expression were observed in patients with weight loss (5.40-fold) and patients without weight loss (5.14-fold) (p=0.04). Patients with T2DM who experienced fatigue showed a 5.49-fold change in expression of the lncRNA NEAT1, while patients with T2DM who did not report fatigue showed a 5.09-fold change in expression of the lncRNA NEAT1 (p=0.01). Patients with T2DM with normal wound healing showed a 4.94-fold change in expression of the lncRNA NEAT1, while patients with reduced or no wound healing showed a 5.74-fold change in expression of the lncRNA NEAT1, and these values were found to be significantly different (p=0.002).
Association of expression of the circulating lncRNA NEAT1 with different parameters among patients with T2DM
Compared with healthy subjects, patients with T2DM with blurred vision showed a 5.53-fold change in expression of the lncRNA NEAT1, while patients with no blurred vision showed a 5.15-fold change in expression of the lncRNA NEAT1 (p=0.008). Patients with T2DM with appetite loss showed a 5.78-fold change in expression of the lncRNA NEAT1, while patients with no appetite loss showed a 4.86-fold change in expression of the lncRNA NEAT1 (p=0.007). Patients with T2DM who smoked showed a 6.41-fold change in expression of the lncRNA NEAT1, while non-smoking patients with T2DM showed a 4.32-fold change in expression of the lncRNA NEAT1, and these values were found to be significantly different (p<0.0001). Compared with healthy subjects, patients with T2DM with alcoholism showed a 6.51-fold change in expression of the lncRNA NEAT1, while patients with T2DM without alcoholism showed a fourfold change in expression of the lncRNA NEAT1, and these values were found to be significantly different (p<0.0001).
Expression of the lncRNA MALAT1 in patients with T2DM
Compared with healthy controls, overall, patients with T2DM showed a 4.46-fold higher expression of the lncRNA MALAT1 (table 3).
Association of expression of the circulating lncRNA MALAT1 with different parameters among patients with T2DM
Among the patients with T2DM, a significant difference in expression of the lncRNA MALAT1 was observed between patients with weight loss (4.85-fold) and patients without weight loss (4.02-fold) (p=0.003). Among the patients with T2DM, patients with blurred vision showed a 5.02-fold change in expression of the lncRNA MALAT1, while patients without blurred vision showed a 4.18-fold change in expression of the lncRNA MALAT1 (p=0.01). Patients with T2DM who smoked showed a 5.09-fold change in expression of the lncRNA MALAT1, while non-smoking patients with T2DM showed a 3.92-fold change in expression of the lncRNA MALAT1, and these values were found to be significantly different (p<0.0001). Compared with healthy subjects, patients with T2DM with alcoholism showed a 5.20-fold change in expression of the lncRNA MALAT1, while patients with T2DM without alcoholism showed a 3.69-fold change in expression of the lncRNA MALAT1, and these values were found to be significantly different (p<0.0001).
Expression of the lncRNA MIAT in patients with T2DM
Compared with healthy subjects, overall, patients with T2DM showed a 6.35-fold higher expression of the lncRNA MIAT (table 4). Patients with T2DM with blurred vision showed a 5.32-fold change in expression of the lncRNA MIAT, while patients without blurred vision showed a 3.72-fold change in expression of the lncRNA MIAT (p<0.0001). Compared with healthy subjects, smoking patients with T2DM showed a 5.41-fold change in expression of the lncRNA MIAT, while non-smoking patients with T2DM showed a 3.27-fold change in expression of the lncRNA MIAT, and these values were found to be significantly different (p<0.0001). Compared with healthy subjects, patients with T2DM with alcoholism showed a 5.74-fold change in expression of the lncRNA MIAT, while patients with T2DM without alcoholism showed a 2.71-fold change in expression of the lncRNA MIAT, and the difference between these values was found to be significant (p<0.0001).
Association of expression of the circulating lncRNA MIAT with different parameters among patients with T2DM
Correlation of the lncRNA NKILA with the lncRNAs NEAT1, MALAT1, and MIAT
A positive correlation was observed between the lncRNA NKILA with NEAT1, MALAT1, and MIAT. Expression of the lncRNA NKILA showed a positive correlation with expression of the lncRNAs NEAT1 (r=0.42, p<0.0001), MALAT1 (r=0.36, p<0.0001) and MIAT (r=0.42, p<0.0001) (figure 1).
Correlation of expression of the lncRNA NKILA with that of the lncRNAs NEAT1, MALAT1, and MIAT among patients with type 1 diabetes mellitus. lncRNA, long non-coding RNA; MALAT1, metastasis-associated lung adenocarcinoma transcript 1; MIAT, myocardial infarction-associated transcript; NEAT1, nuclear enriched abundant transcript 1; NKILA, NF-kappaB interacting lncRNA.
Correlation of the lncRNA NEAT1 with MALAT1 and MIAT
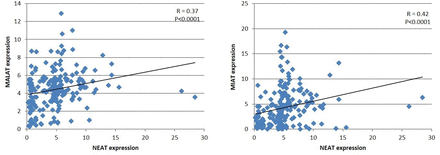
The lncRNA NEAT1 was observed to be positively correlated with MALAT1 and MIAT. Expression of the lncRNA NEAT showed a positive correlation with the expression of the lncRNAs MALAT1 (r=0.37, p<0.0001) and MIAT (r=0.42, p<0.0001) (figure 2).
Correlation of expression of the lncRNA NEAT with that of the lncRNAs MALAT1 and MIAT among patients with type 1 diabetes mellitus. lncRNA, long non-coding RNA; MALAT1, metastasis-associated lung adenocarcinoma transcript 1; MIAT, myocardial infarction-associated transcript; NEAT, nuclear enriched abundant transcript.
Correlation of the lncRNA MALAT1 with MIAT
The lncRNA MALAT1 showed a positive correlation with MIAT. A positive r value was obtained when the expressions of the lncRNAs MALAT1 and MIAT (r=0.26, p<0.0001) were compared (figure 3).
Correlation of expression of the lncRNAs MALAT1 and MIAT among patients with type 2 diabetes mellitus. lncRNA, long non-coding RNA; MALAT1, metastasis-associated lung adenocarcinoma transcript 1; MIAT, myocardial infarction-associated transcript.
Prognostic importance of lncRNA NKILA, NEAT1, MALAT1, MIAT with respect to
T2DM nephropathy and T2DM retinopathy
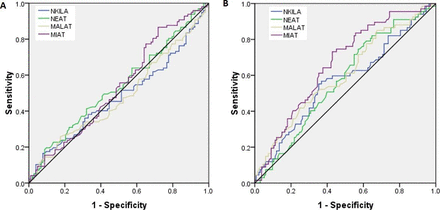
To examine the prognostic significance of lncRNA NKILA, NEAT1, MALAT1, and MIAT in patients with T2DM, two groups were made: patients with nephropathy and without nephropathy, and the receiver operating characteristic (ROC) curve analysis was made (figure 4A). With respect to patients with nephropathy versus patients without nephropathy, the ROC curve of patients with T2DM showed a possible cut-off value of 3.0-fold increase for NKILA, 4.5-fold increase for NEAT1, 4.2-fold increase for MALAT1 and 4.5-fold increase for MIAT; the sensitivity was 54%–52% and 53% and 55%, and the specificity was 42%–48% and 40% and 50%, respectively (AUC=0.48, p=0.63; AUC=0.52, p=0.37; AUC=0.48, p=0.65; and AUC=0.53, p=0.40) (table 5).
ROC curve for lncRNA NKILA, NEAT1, MALAT1, and MIAT (A) with respect to patients with T2DM with nephropathy versus without nephropathy (B) with respect to patients with T2DM with retinopathy versus without retinopathy. lncRNA, long non-coding RNA; MALAT1, metastasis-associated lung adenocarcinoma transcript 1; MIAT, myocardial infarction-associated transcript; NEAT1, nuclear enriched abundant transcript 1; NKILA, NF-kappaB interacting lncRNA; ROC, receiver operating characteristic; T2DM, type 2 diabetes mellitus.
AUC curve for lncRNA NKILA, NEAT1, MALAT1, MIAT with respect to patients with T2DM with nephropathy versus patients without nephropathy and patients with T2DM with retinopathy versus without retinopathy
To examine the prognostic significance of lncRNA NKILA, NEAT1, MALAT1, and MIAT in patients with T2DM, two groups were made: patients with retinopathy and without retinopathy, and the ROC curve analysis was made (figure 4B). The ROC curve, with respect to patients with retinopathy versus patients without retinopathy, of patients with T2DM showed a possible cut-off value of 3.1-fold increase for NKILA, 4.6-fold increase for NEAT1, 4.3-fold increase for MALAT1 and 3.5-fold increase for MIAT and sensitivity was 59%–61% 61% and 68% and specificity was 41%, 51%, 50% and 58%, respectively (AUC=0.56, p=0.12; AUC=0.57, p=0.09; AUC=0.60, p=0.01; AUC=0.66, p<0.0001) (table 5).
Discussion
International Diabetes Federation Atlas22 revealed that approximately 425 million individuals worldwide have diabetes; among them, >90% had T2DM, a complex, multifactorial disease with genetic, epigenetic, and environmental machinery. Several studies have suggested that accelerated aging, cellular senescence, and proinflammatory mechanism are closely linked to the etiology of type 2 diabetes and insulin resistance.23 Epigenetic changes and alterations have been a major part in the inflammation regulation and cellular senescence frequently linked with type 2 diabetes.24 lncRNAs have progressively came into consideration due to increased recognition of their functional significance in health and diseases.8 lncRNAs have been also established afterward to protein-coding genes under the compact transcriptional mechanism, and expression pattern is usually associated with tissue diversity and disease severity.9 The role of lncRNAs been lately recognized in the T2DM pathogenesis and related complications, as well as its involvement in pathophysiological mechanisms linked to the genesis, advancement and the worsening of the disease.25 26
Our present study reported increased expression levels of the lncRNAs NKILA, NEAT1, MALAT1, and MIAT in patients with T2DM. Patients with T2DM showed more than threefold higher expression of the lncRNA NKILA compared with healthy controls. Higher expression levels of the lncRNA NKILA were observed in patients with T2DM with hypertension, smoking patients with T2DM, and patients with T2DM with alcoholism compared with their corresponding counterparts.
A study by Li et al in 201927 demonstrated that lncRNA NKILA was specifically upregulated in patients with T2DM who later developed diabetic-related complications such as cardiomyopathy, suggesting that overexpression of the lncRNA NKILA may contribute to cardiomyocyte apoptosis and its involvement in the progression of diabetic cardiomyopathy (DC). It was suggested that suppressing expression of the lncRNA NKILA may inhibit the development of DC.27
The apoptosis of cardiomyocytes under high-glucose conditions contributes to the pathogenesis of DC,28 overexpression, as well as knockdown of the lncRNA NKILA accelerated and inhibited apoptotic cell death, respectively.27 Compared with healthy subjects, patients with T2DM showed a more than fivefold increase in expression of the lncRNA NEAT1. Patients with T2DM with weight loss and fatigue showed higher lncRNA NEAT1 expression compared with their counterparts; patients with T2DM with wound healing showed higher expression of the lncRNA NEAT1 compared with those with slow wound healing. NEAT1 is also highly expressed due to hyperglycemia and NEAT1 with the AKT/mTOR pathway.29 Recent studies have shown that lnc-NEAT1 is aberrantly expressed in diabetic mice; moreover, some studies have shown that lnc-NEAT1 has proinflammatory influence through affecting inflammatory pathways, such as its activation of mitogen-activated protein kinase (MAPK) pathways or the toll-like receptor 3–p38 pathway in some inflammatory diseases.5 30 A study by Ma et al reported that high glucose levels are positively associated with overexpression of the lncRNA NEAT1, indicating that lnc-NEAT1 was overexpressed in the cellular model of DN.31 Overexpression of the lncRNA NEAT1 was also observed in myocardial I/R injury cells compared with normal myocardial cells.32
The lncRNA MALAT1 showed more than fourfold higher expression in patients with T2DM compared with healthy controls. Among patients with T2DM, higher expression of the lncRNA MALAT1 was linked with weight loss, blurred vision, smoking, and alcoholism compared with the corresponding counterparts without these parameters.
MALAT1 is a highly conserved lncRNA that has been related to multiple pathological conditions, including diabetes-related complications.33 Current studies have shown that MALAT1 could play significant roles in the pathophysiological condition, tissue inflammation, and progression of diabetes by modulating gene transcription.34 MALAT1 expression was elevated by high glucose in diabetic cataract tissue cells, provoking apoptosis and oxidative stress.34
Yan et al reported that MALAT1 was considerably upregulated in diabetic mouse retinas, which contributed to the occurrence of diabetic retinopathy in an in vitro animal study, and MALAT1 was also upregulated in clinical samples.35 MALAT1 overexpression serves as essential pathogenic machinery for diabetes-related dysfunction and endothelial cell proliferation through p38MAPK signaling. MALAT1 reticence may become a potent antiangiogenic therapy for diabetic microvascular difficulties.36
Zhang et al explored the relationship among the lncRNAs MALAT1, p21, and H19 and gestational diabetes mellitus (GDM) and reported the lncRNA MALAT1 expression level was significantly higher in the GDM group compared with its counterpart. Moreover, the lncRNA MALAT1 is inter-related with the expression of the lncRNAs p21 and H19. MALAT1 was acknowledged as a unique serum biomarker that predicts GDM. Detection of MALAT1 expression provides a promising biomarker for future strategies to diagnose GDM, and GDM may be treatable by regulating the expression of the lncRNA MALAT1.37
MIAT was one of the most highly expressed (>6 fold) lncRNAs in patients with T2DM compared with healthy controls. Its high expression was observed in patients with T2DM with blurred vision, and MIAT may be a leading factor in diabetic-related eye disorders. High MIAT expression was also observed to be associated with smoking and alcoholism among patients with T2DM.
Researchers have shown that the unusual expression of MIAT has participated in cell death, and various diseases development, such as myocardial infarction,38 microvascular dysfunction,39 and diabetes.40 An experiment in mice by Zhang et al in 2017 suggested that streptozotocin (STZ) supplementation significantly enhances the MIAT expression level, and highglucose supplementation promoted expression of the lncRNA MIAT.41
MIAT is particularly involved in diabetic retinopathy and numerous other microvascular problems.42 A study by Toraih et al in 201943 revealed a significantly higher expression of circulating MIAT in patients with diabetes with coronary heart disease (CAD). Higher expression levels of MIAT and MALAT1 were related to hypertension and premature CAD, and both lncRNAs displayed greater relative expression in patients with a progressive history of prior cardiac ischaemic events.
Altered MALAT1 expression was reported to be closely related to diabetic complications in previous studies.44 Moreover, the greater relative expression of MALAT1 observed in diabetic and hypertensive subgroups of patients with CAD suggests a possible role by which diabetes and hypertension produce endothelial alteration and accelerate the atherosclerosis rate.45 Recent studies reported the potential contributions of MIAT and MALAT1 to endothelial dysfunction in patients with diabetes.46 This study analyzed the prognostic importance of circulating lnc NKILA, NEAT1, MALAT1, and MIAT in patients with T2DM and observed that the MALAT1 and MEAT could be used as prognostic markers for patients with T2DM with retinopathy.
Conclusion
The present study reports the clinical relevance of the increased expression of circulating NKILA, NEAT1, MALAT, and MIAT in patients with T2DM with poor patient outcomes. All four lncRNAs were significantly linked with alcoholism, and smoking may influence the degree and severity of disease among patients with T2DM. Overexpression of the lncRNAs NKILA, NEAT1, MALAT, and MIAT showed a positive correlation, and together, these lncRNAs may contribute to the progression of T2DM or contribute to other related diabetes-related complications. A better understanding of the underlying functions and mechanisms of lncRNAs will assist us to recognise the pathophysiology of diabetes and its related difficulties and thereby to use these to adapt treatment strategies and to develop novel therapeutic approaches in the near future.
Acknowledgments
The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University for funding this work through the General Research Project under grant number (G.R.P- 298).
References
Supplementary materials
Supplementary Data
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
Supplementary Data
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
Footnotes
MA and MMAB contributed equally.
Correction notice This article has been corrected since it was published. Author name has been corrected to Mohammad Yahya Alshahrani.
Contributors Conceptualization: MA, MMAB and AKV; methodology: MA, MYA and IA; software: AGA and MMAB; validation: PCJ; formal analysis: MMAB, AKV, OMA, and AMA; data curation: MMAB and MA; writing (original draft preparation): MMAB and AKV; writing (review and editing): MA, MMAB, MYA, AGA, and AKV; visualization: PCJ; supervision: MA.
Funding The Deanship of Scientific Research at King Khalid University funded this work through the General Research Project under grant number (G.R.P- 298).
Competing interests None declared.
Patient consent for publication Not required.
Ethics approval The study has been conducted only after the due clearance and approval from the ethics committee of GKV (vide proposal number 14/08/2015/GKV/IEC/2015) has been obtained. As part of a mandatory standardized ethical norm, written with signed informed consent was taken from the particular person before inclusion in the research work.
Provenance and peer review Not commissioned; externally peer reviewed.
Data availability statement Data may be obtained from a third party and are not publicly available. The datasets used and analyzed during the present study are available from the corresponding author.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.